+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
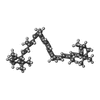
| Title | Structure of PSII-ACPII supercomplex from cryptophyte algae | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Photosystem II / PHOTOSYNTHESIS | |||||||||
| Function / homology |  Function and homology information Function and homology informationthylakoid membrane / photosystem II stabilization / photosystem II reaction center / photosystem II / oxidoreductase activity, acting on diphenols and related substances as donors, oxygen as acceptor / photosynthetic electron transport chain / photosystem II / response to herbicide / plastid / chlorophyll binding ...thylakoid membrane / photosystem II stabilization / photosystem II reaction center / photosystem II / oxidoreductase activity, acting on diphenols and related substances as donors, oxygen as acceptor / photosynthetic electron transport chain / photosystem II / response to herbicide / plastid / chlorophyll binding / photosynthetic electron transport in photosystem II / phosphate ion binding / photosynthesis, light reaction / chloroplast thylakoid membrane / photosynthesis / chloroplast / respiratory electron transport chain / oxidoreductase activity / electron transfer activity / protein stabilization / iron ion binding / heme binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Chroomonas placoidea (eukaryote) Chroomonas placoidea (eukaryote) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.47 Å | |||||||||
 Authors Authors | Li XY / Mao ZY / Shen JR / Han GY | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structure and distinct supramolecular organization of a PSII-ACPII dimer from a cryptophyte alga Chroomonas placoidea. Authors: Zhiyuan Mao / Xingyue Li / Zhenhua Li / Liangliang Shen / Xiaoyi Li / Yanyan Yang / Wenda Wang / Tingyun Kuang / Jian-Ren Shen / Guangye Han /   Abstract: Cryptophyte algae are an evolutionarily distinct and ecologically important group of photosynthetic unicellular eukaryotes. Photosystem II (PSII) of cryptophyte algae associates with alloxanthin ...Cryptophyte algae are an evolutionarily distinct and ecologically important group of photosynthetic unicellular eukaryotes. Photosystem II (PSII) of cryptophyte algae associates with alloxanthin chlorophyll a/c-binding proteins (ACPs) to act as the peripheral light-harvesting system, whose supramolecular organization is unknown. Here, we purify the PSII-ACPII supercomplex from a cryptophyte alga Chroomonas placoidea (C. placoidea), and analyze its structure at a resolution of 2.47 Å using cryo-electron microscopy. This structure reveals a dimeric organization of PSII-ACPII containing two PSII core monomers flanked by six symmetrically arranged ACPII subunits. The PSII core is conserved whereas the organization of ACPII subunits exhibits a distinct pattern, different from those observed so far in PSII of other algae and higher plants. Furthermore, we find a Chl a-binding antenna subunit, CCPII-S, which mediates interaction of ACPII with the PSII core. These results provide a structural basis for the assembly of antennas within the supercomplex and possible excitation energy transfer pathways in cryptophyte algal PSII, shedding light on the diversity of supramolecular organization of photosynthetic machinery. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37414.map.gz emd_37414.map.gz | 257.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37414-v30.xml emd-37414-v30.xml emd-37414.xml emd-37414.xml | 48.6 KB 48.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37414_fsc.xml emd_37414_fsc.xml | 16.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_37414.png emd_37414.png | 74.8 KB | ||
| Filedesc metadata |  emd-37414.cif.gz emd-37414.cif.gz | 10.5 KB | ||
| Others |  emd_37414_half_map_1.map.gz emd_37414_half_map_1.map.gz emd_37414_half_map_2.map.gz emd_37414_half_map_2.map.gz | 475.4 MB 475.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37414 http://ftp.pdbj.org/pub/emdb/structures/EMD-37414 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37414 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37414 | HTTPS FTP |
-Related structure data
| Related structure data |  8wb4MC  8xklC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37414.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37414.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
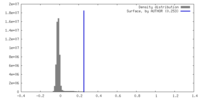
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_37414_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
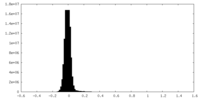
| Density Histograms |
-Half map: #1
| File | emd_37414_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
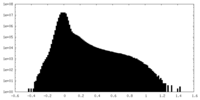
| Density Histograms |
- Sample components
Sample components
+Entire : Structure of PSII-ACPII supercomplex from cryptophyte algae
+Supramolecule #1: Structure of PSII-ACPII supercomplex from cryptophyte algae
+Macromolecule #1: Photosystem II reaction center M
+Macromolecule #2: ACPII-1
+Macromolecule #3: ACPII-2
+Macromolecule #4: ACPII-3
+Macromolecule #5: ACPII-4
+Macromolecule #6: ACPII-5
+Macromolecule #7: ACPII-6
+Macromolecule #8: Photosystem II protein D1
+Macromolecule #9: Photosystem II D2 protein
+Macromolecule #10: Cytochrome b559 subunit alpha
+Macromolecule #11: Cytochrome b559 subunit beta
+Macromolecule #12: Photosystem II reaction center protein H
+Macromolecule #13: Photosystem II reaction center protein I
+Macromolecule #14: Photosystem II reaction center protein J
+Macromolecule #15: Photosystem II reaction center protein K
+Macromolecule #16: Photosystem II reaction center protein O
+Macromolecule #17: Photosystem II reaction center protein Q
+Macromolecule #18: Photosystem II reaction center protein R
+Macromolecule #19: CCPII-S
+Macromolecule #20: Photosystem II reaction center protein T
+Macromolecule #21: Photosystem II reaction center protein U
+Macromolecule #22: Photosystem II cytochrome c550
+Macromolecule #23: Photosystem II reaction center X protein
+Macromolecule #24: Photosystem II reaction center protein Ycf12
+Macromolecule #25: Photosystem II reaction center protein Z
+Macromolecule #26: Photosystem II CP47 reaction center protein
+Macromolecule #27: Photosystem II CP43 reaction center protein
+Macromolecule #28: Photosystem II reaction center protein G
+Macromolecule #29: Photosystem II reaction center protein L
+Macromolecule #30: Photosystem II reaction center protein W
+Macromolecule #31: (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta...
+Macromolecule #32: CHLOROPHYLL A
+Macromolecule #33: Chlorophyll c2
+Macromolecule #34: (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E}...
+Macromolecule #35: (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E}...
+Macromolecule #36: (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E}...
+Macromolecule #37: 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE
+Macromolecule #38: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE
+Macromolecule #39: 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL
+Macromolecule #40: CA-MN4-O5 CLUSTER
+Macromolecule #41: PHEOPHYTIN A
+Macromolecule #42: CHLORIDE ION
+Macromolecule #43: 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,...
+Macromolecule #44: BICARBONATE ION
+Macromolecule #45: DIGALACTOSYL DIACYL GLYCEROL (DGDG)
+Macromolecule #46: FE (II) ION
+Macromolecule #47: PROTOPORPHYRIN IX CONTAINING FE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)