[English] 日本語
 Yorodumi
Yorodumi- EMDB-37227: Structure of African swine fever virus topoisomerase II in comple... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of African swine fever virus topoisomerase II in complex with dsDNA | |||||||||
 Map data Map data | C | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | topo 2 / VIRAL PROTEIN-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsister chromatid segregation / DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) activity / DNA topoisomerase (ATP-hydrolysing) / DNA topological change / host cell cytoplasm / DNA binding / ATP binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |   African swine fever virus / DNA molecule (others) African swine fever virus / DNA molecule (others) | |||||||||
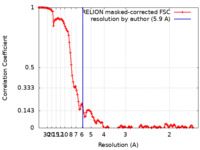
| Method | single particle reconstruction / cryo EM / Resolution: 5.9 Å | |||||||||
 Authors Authors | Cong J / Xin Y / Li X / Chen Y | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural insights into the DNA topoisomerase II of the African swine fever virus. Authors: Jingyuan Cong / Yuhui Xin / Huiling Kang / Yunge Yang / Chenlong Wang / Dongming Zhao / Xuemei Li / Zihe Rao / Yutao Chen /  Abstract: Type II topoisomerases are ubiquitous enzymes that play a pivotal role in modulating the topological configuration of double-stranded DNA. These topoisomerases are required for DNA metabolism and ...Type II topoisomerases are ubiquitous enzymes that play a pivotal role in modulating the topological configuration of double-stranded DNA. These topoisomerases are required for DNA metabolism and have been extensively studied in both prokaryotic and eukaryotic organisms. However, our understanding of virus-encoded type II topoisomerases remains limited. One intriguing example is the African swine fever virus, which stands as the sole mammalian-infecting virus encoding a type II topoisomerase. In this work, we use several approaches including cryo-EM, X-ray crystallography, and biochemical assays to investigate the structure and function of the African swine fever virus type II topoisomerase, pP1192R. We determine the structures of pP1192R in different conformational states and confirm its enzymatic activity in vitro. Collectively, our results illustrate the basic mechanisms of viral type II topoisomerases, increasing our understanding of these enzymes and presenting a potential avenue for intervention strategies to mitigate the impact of the African swine fever virus. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37227.map.gz emd_37227.map.gz | 161.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37227-v30.xml emd-37227-v30.xml emd-37227.xml emd-37227.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37227_fsc.xml emd_37227_fsc.xml | 12.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_37227.png emd_37227.png | 26.7 KB | ||
| Filedesc metadata |  emd-37227.cif.gz emd-37227.cif.gz | 6.7 KB | ||
| Others |  emd_37227_half_map_1.map.gz emd_37227_half_map_1.map.gz emd_37227_half_map_2.map.gz emd_37227_half_map_2.map.gz | 140.9 MB 141 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37227 http://ftp.pdbj.org/pub/emdb/structures/EMD-37227 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37227 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37227 | HTTPS FTP |
-Related structure data
| Related structure data |  8kgnMC  8kglC  8kgmC  8kgoC  8kgpC  8kgqC  8kgrC  8kgsC  8kgtC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37227.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37227.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | C | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: C
| File | emd_37227_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | C | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: C
| File | emd_37227_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | C | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : pP1192R
| Entire | Name: pP1192R |
|---|---|
| Components |
|
-Supramolecule #1: pP1192R
| Supramolecule | Name: pP1192R / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: dimer |
|---|---|
| Source (natural) | Organism:   African swine fever virus African swine fever virus |
-Macromolecule #1: DNA topoisomerase 2
| Macromolecule | Name: DNA topoisomerase 2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   African swine fever virus African swine fever virus |
| Molecular weight | Theoretical: 138.093359 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EFATMEAFEI SDFKEHAKKK SMWAGALNKV TISGLMGVFT EDEDLMALPI HRDHCPALLK IFDEIIVNAT DHERACHNKT KKVTYIKIS FDKGVFSCEN DGPGIPIAKH EQASLIAKRD VYVPEVASCH FLAGTNINKA KDCIKGGTNG VGLKLAMVHS Q WAILTTAD ...String: EFATMEAFEI SDFKEHAKKK SMWAGALNKV TISGLMGVFT EDEDLMALPI HRDHCPALLK IFDEIIVNAT DHERACHNKT KKVTYIKIS FDKGVFSCEN DGPGIPIAKH EQASLIAKRD VYVPEVASCH FLAGTNINKA KDCIKGGTNG VGLKLAMVHS Q WAILTTAD GAQKYVQHIN QRLDIIEPPT ITPSREMFTR IELMPVYQEL GYAEPLSETE QADLSAWIYL RACQCAAYVG KG TTIYYND KPCRTGSVMA LAKMYTLLSA PNSTIHTATI KADAKPYSLH PLQVAAVVSP KFKKFEHVSV INGVNCVKGE HVT FLKKTI NEMVVKKFQQ TIKDKNRKTT LRDSCSNIFI VIVGSIPGIE WTGQRKDELS IAENVFKTHY SIPSSFLTSM TKSI VDILL QSISKKDNHK QVDVDKYTRA RNAGGKRAQD CMLLAAEGDS ALSLLRTGLT LGKSNPSGPS FDFCGMISLG GVIMN ACKK VTNITTDSGE TIMVRNEQLT NNKVLQGIVQ VLGLDFNCHY KTQEERAKLR YGCIVACVDQ DLDGCGKILG LLLAYF HLF WPQLIIHGFV KRLLTPLIRV YEKGKTMPVE FYYEQEFDAW AKKQTSLANH TVKYYKGLAA HDTHEVKSMF KHFDNMV YT FTLDDSAKEL FHIYFGGESE LRKRELCTGV VPLTETQTQS IHSVRRIPCS LHLQVDTKAY KLDAIERQIP NFLDGMTR A RRKILAGGVK CFASNNRERK VFQFGGYVAD HMFYHHGDMS LNTSIIKAAQ YYPGSSHLYP VFIGIGSFGS RHLGGKDAG SPRYISVQLA SEFIKTMFPA EDSWLLPYVF EDGQRAEPEY YVPVLPLAIM EYGANPSEGW KYTTWARQLE DILALVRAYV DKDNPKHEL LHYAIKHKIT ILPLRPSNYN FKGHLKRFGQ YYYSYGTYVI SEQRNIITIT ELPLRVPTVA YIESIKKSSN R MTFIEEII DYSSSETIEI LVKLKPNSLN RIVEEFKETE EQDSIENFLR LRNCLHSHLN FVKPKGGIIE FNTYYEILYA WL PYRRELY QKRLMREHAV LKLRIIMETA IVRYINESAE LNLSHYEDEK EASRILSEHG FPPLNHTLII SPEFASIEEL NQK ALQGCY TYILSLQARE LLIAAKTRRV EKIKKMQARL DKVEQLLQES PFPGASVWLE EIDAVEKAII KGRNTQWKFH ENLY FQGHH HHHHHH UniProtKB: DNA topoisomerase 2 |
-Macromolecule #2: DNA (38-MER)
| Macromolecule | Name: DNA (38-MER) / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: DNA molecule (others) |
| Molecular weight | Theoretical: 16.058391 KDa |
| Sequence | String: (DA)(DT)(DG)(DC)(DA)(DT)(DA)(DT)(DA)(DT) (DA)(DT)(DG)(DT)(DA)(DT)(DA)(DT)(DG)(DT) (DA)(DT)(DG)(DT)(DG)(DT)(DG)(DT)(DA) (DT)(DA)(DT)(DA)(DT)(DA)(DC)(DA)(DC)(DA) (DT) (DA)(DT)(DA)(DT)(DA)(DT) ...String: (DA)(DT)(DG)(DC)(DA)(DT)(DA)(DT)(DA)(DT) (DA)(DT)(DG)(DT)(DA)(DT)(DA)(DT)(DG)(DT) (DA)(DT)(DG)(DT)(DG)(DT)(DG)(DT)(DA) (DT)(DA)(DT)(DA)(DT)(DA)(DC)(DA)(DC)(DA) (DT) (DA)(DT)(DA)(DT)(DA)(DT)(DA)(DT) (DA)(DT)(DA)(DT) |
-Macromolecule #3: DNA (38-MER)
| Macromolecule | Name: DNA (38-MER) / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: DNA molecule (others) |
| Molecular weight | Theoretical: 15.965361 KDa |
| Sequence | String: (DA)(DT)(DA)(DT)(DA)(DT)(DA)(DT)(DA)(DT) (DA)(DT)(DA)(DT)(DG)(DT)(DG)(DT)(DA)(DT) (DA)(DT)(DA)(DT)(DA)(DC)(DA)(DC)(DA) (DC)(DA)(DT)(DA)(DC)(DA)(DT)(DA)(DT)(DA) (DC) (DA)(DT)(DA)(DT)(DA)(DT) ...String: (DA)(DT)(DA)(DT)(DA)(DT)(DA)(DT)(DA)(DT) (DA)(DT)(DA)(DT)(DG)(DT)(DG)(DT)(DA)(DT) (DA)(DT)(DA)(DT)(DA)(DC)(DA)(DC)(DA) (DC)(DA)(DT)(DA)(DC)(DA)(DT)(DA)(DT)(DA) (DC) (DA)(DT)(DA)(DT)(DA)(DT)(DA)(DT) (DG)(DC)(DA)(DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)