[English] 日本語
 Yorodumi
Yorodumi- EMDB-3701: Structure of Rubisco from Rhodobacter sphaeroides in complex with... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3701 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Rubisco from Rhodobacter sphaeroides in complex with CABP and RcaCC | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Rhodobacter sphaeroides (bacteria) Rhodobacter sphaeroides (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.56 Å | |||||||||
 Authors Authors | Bracher A / Milicic G / Ciniawsky S / Wendler P / Hayer-Hartl M / Hartl FU | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2017 Journal: Mol Cell / Year: 2017Title: Mechanism of Enzyme Repair by the AAA Chaperone Rubisco Activase. Authors: Javaid Y Bhat / Goran Miličić / Gabriel Thieulin-Pardo / Andreas Bracher / Andrew Maxwell / Susanne Ciniawsky / Oliver Mueller-Cajar / John R Engen / F Ulrich Hartl / Petra Wendler / Manajit Hayer-Hartl /   Abstract: How AAA+ chaperones conformationally remodel specific target proteins in an ATP-dependent manner is not well understood. Here, we investigated the mechanism of the AAA+ protein Rubisco activase (Rca) ...How AAA+ chaperones conformationally remodel specific target proteins in an ATP-dependent manner is not well understood. Here, we investigated the mechanism of the AAA+ protein Rubisco activase (Rca) in metabolic repair of the photosynthetic enzyme Rubisco, a complex of eight large (RbcL) and eight small (RbcS) subunits containing eight catalytic sites. Rubisco is prone to inhibition by tight-binding sugar phosphates, whose removal is catalyzed by Rca. We engineered a stable Rca hexamer ring and analyzed its functional interaction with Rubisco. Hydrogen/deuterium exchange and chemical crosslinking showed that Rca structurally destabilizes elements of the Rubisco active site with remarkable selectivity. Cryo-electron microscopy revealed that Rca docks onto Rubisco over one active site at a time, positioning the C-terminal strand of RbcL, which stabilizes the catalytic center, for access to the Rca hexamer pore. The pulling force of Rca is fine-tuned to avoid global destabilization and allow for precise enzyme repair. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3701.map.gz emd_3701.map.gz | 20.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3701-v30.xml emd-3701-v30.xml emd-3701.xml emd-3701.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3701.png emd_3701.png | 70.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3701 http://ftp.pdbj.org/pub/emdb/structures/EMD-3701 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3701 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3701 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3701.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3701.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
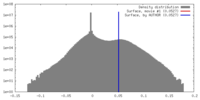
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Rubisco-RcaCC complex
| Entire | Name: Rubisco-RcaCC complex |
|---|---|
| Components |
|
-Supramolecule #1: Rubisco-RcaCC complex
| Supramolecule | Name: Rubisco-RcaCC complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Rubisco was treated with the inhibitor CABP. RcaCC, ATP, ATP-gammaS and RuBP were added. The sample was treated with glutaraldehyde crosslinker. Rubisco-RcaCC complexes were purified by SEC. |
|---|---|
| Source (natural) | Organism:  Rhodobacter sphaeroides (bacteria) Rhodobacter sphaeroides (bacteria) |
| Recombinant expression | Organism:  |
| Molecular weight | Experimental: 768 KDa |
-Macromolecule #1: CbbX
| Macromolecule | Name: CbbX / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Rhodobacter sphaeroides (bacteria) Rhodobacter sphaeroides (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MTDAATAPTS IDLRAEYEGS GAKEVLEELD RELIGLKPVK DRIRETAAcL LVERARQKLG LAHETPTLH MSFTGNPGTG KTTVALKMAG LLHRLGYVRK GHLVSVTRDD LVGQYIGHTA P KTKEVLKR AMGGVLFIDE AYYLYRPDNE RDYGQEAIEI LLQVMENNRD ...String: MTDAATAPTS IDLRAEYEGS GAKEVLEELD RELIGLKPVK DRIRETAAcL LVERARQKLG LAHETPTLH MSFTGNPGTG KTTVALKMAG LLHRLGYVRK GHLVSVTRDD LVGQYIGHTA P KTKEVLKR AMGGVLFIDE AYYLYRPDNE RDYGQEAIEI LLQVMENNRD DLVVILAGYA DR MENFFQS NPGFRSRIAH HIEFPDYSDE ELFEIAGHML DDQNYQMTPE AETALRAYIG LRR NQPHFA NARSIRNALD RARLRQcNRL FTASSGPLDA RALSTMAEED IRASRVFKGG LDSE RRAAE ALAR |
-Macromolecule #2: RbcL
| Macromolecule | Name: RbcL / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Rhodobacter sphaeroides (bacteria) Rhodobacter sphaeroides (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MDTKTTEIKG KERYKAGVLK YAQMGYWDGD YVPKDTDVLA LFRITPQEGV DPVEAAAAVA GESSTATWT VVWTDRLTAC DSYRAKAYRV EPVPGTPGQY FCYVAYDLIL FEEGSIANLT A SIIGNVFS FKPLKAARLE DMRFPVAYVK TYKGPPTGIV GERERLDKFG ...String: MDTKTTEIKG KERYKAGVLK YAQMGYWDGD YVPKDTDVLA LFRITPQEGV DPVEAAAAVA GESSTATWT VVWTDRLTAC DSYRAKAYRV EPVPGTPGQY FCYVAYDLIL FEEGSIANLT A SIIGNVFS FKPLKAARLE DMRFPVAYVK TYKGPPTGIV GERERLDKFG KPLLGATTKP KL GLSGKNY GRVVYEGLKG GLDFMKDDEN INSQPFMHWR DRFLYVMEAV NLASAQTGEV KGH YLNITA GTMEEMYRRA EFAKSLGSVI VMVDLIIGYT AIQSISEWCR QNDMILHMHR AGHG TYTRQ KNHGISFRVI AKWLRLAGVD HLHCGTAVGK LEGDPLTVQG YYNVCREPFN TVDLP RGIF FEQDWADLRK VMPVASGGIH AGQMHQLLSL FGDDVVLQFG GGTIGHPMGI QAGATA NRV ALEAMVLARN EGRNIDVEGP EILRAAAKWC KPLEAALDTW GNITFNYTST DTSDFVP TA SVAM |
-Macromolecule #3: RbcS
| Macromolecule | Name: RbcS / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Rhodobacter sphaeroides (bacteria) Rhodobacter sphaeroides (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MRITQGCFSF LPDLTDEQIS AQVDYCLGRG WAVSLEHTDD PHPRNTYWEM WGMPMFDLRD PKGVMIELDE CRKAWPGRYI RINAFDSTRG FETVTMSFIV NRPEVEPSLR MERTEVDGRS IRYTHSIVR |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Support film - Material: CARBON / Support film - Film thickness: 2.0 nm / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV | ||||||||||||||||||
| Details | RcaCC hexamers (20 micromolar monomer) were mixed with E.C.M-CABP octamers (10 micromolar monomer) in a reaction containing 20 mM HEPES pH 7.5, 50 mM NaCl, 10 mM MgCl2, 10 mM ATP and 1mM RuBP, for 1 min at 25oC prior to addition of 0.125 % of glutaraldehyde (GA). After 10 min the reaction was quenched by addition of 0.1M Tris HCl pH 8 followed by gel filtration on a Superdex 200 PC 3.2/30 column (GE Healthcare).The fractions were eluted in buffer A and analyzed on a 6 % native gel. Fraction 13 containing HMW complexes with the least amount of free Rubisco were chosen for cryo-EM. The crosslinked E.C.M.-CABP-RcaCC complexes were diluted to 0.0030-0.0035 g ml-1 in 20 mM Tris-HCl pH 8.0, 50 mM NaCl, 1 mM ATP, 1 mM ATP-gammaS and 1 mM RuBP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Details | Cs corrected Krios 1 at NeCEN (June 2016) |
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average exposure time: 1.25 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Software - Name: CTFFIND (ver. 4) |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 7.56 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION / Number images used: 333711 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: OTHER / Software - Name: RELION |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)