+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human TRPV4 with antagonist GSK279 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Channel / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationstretch-activated, monoatomic cation-selective, calcium channel activity / blood vessel endothelial cell delamination / regulation of response to osmotic stress / osmosensor activity / vasopressin secretion / calcium ion import into cytosol / negative regulation of brown fat cell differentiation / positive regulation of microtubule depolymerization / positive regulation of striated muscle contraction / positive regulation of macrophage inflammatory protein 1 alpha production ...stretch-activated, monoatomic cation-selective, calcium channel activity / blood vessel endothelial cell delamination / regulation of response to osmotic stress / osmosensor activity / vasopressin secretion / calcium ion import into cytosol / negative regulation of brown fat cell differentiation / positive regulation of microtubule depolymerization / positive regulation of striated muscle contraction / positive regulation of macrophage inflammatory protein 1 alpha production / hyperosmotic salinity response / positive regulation of chemokine (C-X-C motif) ligand 1 production / positive regulation of chemokine (C-C motif) ligand 5 production / cartilage development involved in endochondral bone morphogenesis / cellular hypotonic salinity response / cellular hypotonic response / cortical microtubule organization / multicellular organismal-level water homeostasis / positive regulation of vascular permeability / osmosensory signaling pathway / cell-cell junction assembly / positive regulation of monocyte chemotactic protein-1 production / calcium ion import / cell volume homeostasis / cellular response to osmotic stress / regulation of aerobic respiration / TRP channels / cortical actin cytoskeleton / diet induced thermogenesis / positive regulation of macrophage chemotaxis / microtubule polymerization / calcium ion import across plasma membrane / response to mechanical stimulus / alpha-tubulin binding / beta-tubulin binding / monoatomic cation channel activity / cytoplasmic microtubule / SH2 domain binding / actin filament organization / protein kinase C binding / filopodium / adherens junction / positive regulation of JNK cascade / response to insulin / calcium ion transmembrane transport / positive regulation of interleukin-6 production / calcium channel activity / ruffle membrane / intracellular calcium ion homeostasis / positive regulation of inflammatory response / actin filament binding / calcium ion transport / glucose homeostasis / lamellipodium / negative regulation of neuron projection development / cellular response to heat / positive regulation of cytosolic calcium ion concentration / growth cone / actin binding / actin cytoskeleton organization / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / microtubule binding / response to hypoxia / calmodulin binding / positive regulation of ERK1 and ERK2 cascade / cilium / apical plasma membrane / focal adhesion / lipid binding / protein kinase binding / cell surface / negative regulation of transcription by RNA polymerase II / endoplasmic reticulum / ATP binding / metal ion binding / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.45 Å | |||||||||
 Authors Authors | Fan J / Lei X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Adv Sci (Weinh) / Year: 2024 Journal: Adv Sci (Weinh) / Year: 2024Title: Structural Pharmacology of TRPV4 Antagonists. Authors: Junping Fan / Chang Guo / Daohong Liao / Han Ke / Jing Lei / Wenjun Xie / Yuliang Tang / Makoto Tominaga / Zhuo Huang / Xiaoguang Lei /   Abstract: The nonselective calcium-permeable Transient Receptor Potential Cation Channel Subfamily V Member4 (TRPV4) channel regulates various physiological activities. Dysfunction of TRPV4 is linked to many ...The nonselective calcium-permeable Transient Receptor Potential Cation Channel Subfamily V Member4 (TRPV4) channel regulates various physiological activities. Dysfunction of TRPV4 is linked to many severe diseases, including edema, pain, gastrointestinal disorders, lung diseases, and inherited neurodegeneration. Emerging TRPV4 antagonists show potential clinical benefits. However, the molecular mechanisms of TRPV4 antagonism remain poorly understood. Here, cryo-electron microscopy (cryo-EM) structures of human TRPV4 are presented in-complex with two potent antagonists, revealing the detailed binding pockets and regulatory mechanisms of TRPV4 gating. Both antagonists bind to the voltage-sensing-like domain (VSLD) and stabilize the channel in closed states. These two antagonists induce TRPV4 to undergo an apparent fourfold to twofold symmetry transition. Moreover, it is demonstrated that one of the antagonists binds to the VSLD extended pocket, which differs from the canonical VSLD pocket. Complemented with functional and molecular dynamics simulation results, this study provides crucial mechanistic insights into TRPV4 regulation by small-molecule antagonists, which may facilitate future drug discovery targeting TRPV4. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36660.map.gz emd_36660.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36660-v30.xml emd-36660-v30.xml emd-36660.xml emd-36660.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36660.png emd_36660.png | 119 KB | ||
| Filedesc metadata |  emd-36660.cif.gz emd-36660.cif.gz | 7 KB | ||
| Others |  emd_36660_half_map_1.map.gz emd_36660_half_map_1.map.gz emd_36660_half_map_2.map.gz emd_36660_half_map_2.map.gz | 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36660 http://ftp.pdbj.org/pub/emdb/structures/EMD-36660 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36660 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36660 | HTTPS FTP |
-Validation report
| Summary document |  emd_36660_validation.pdf.gz emd_36660_validation.pdf.gz | 874 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36660_full_validation.pdf.gz emd_36660_full_validation.pdf.gz | 873.6 KB | Display | |
| Data in XML |  emd_36660_validation.xml.gz emd_36660_validation.xml.gz | 12.2 KB | Display | |
| Data in CIF |  emd_36660_validation.cif.gz emd_36660_validation.cif.gz | 14.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36660 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36660 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36660 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36660 | HTTPS FTP |
-Related structure data
| Related structure data |  8ju6MC  8ju5C  8jviC  8jvjC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36660.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36660.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_36660_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_36660_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : TRPV4
| Entire | Name: TRPV4 |
|---|---|
| Components |
|
-Supramolecule #1: TRPV4
| Supramolecule | Name: TRPV4 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 400 KDa |
-Macromolecule #1: Transient receptor potential cation channel subfamily V member 4,...
| Macromolecule | Name: Transient receptor potential cation channel subfamily V member 4,3C-GFP type: protein_or_peptide / ID: 1 Details: Author stated: The section (872-874) is the cloning site. The domain (875-882) is PreScission Site. The domain (883-1116) is corresponding to this sfGFP (462-695 amino acids, GenBank: ...Details: Author stated: The section (872-874) is the cloning site. The domain (875-882) is PreScission Site. The domain (883-1116) is corresponding to this sfGFP (462-695 amino acids, GenBank: ALP48449.1). The domain (1117-1144) is the expression Tag. Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 128.628547 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MADSSEGPRA GPGEVAELPG DESGTPGGEA FPLSSLANLF EGEDGSLSPS PADASRPAGP GDGRPNLRMK FQGAFRKGVP NPIDLLEST LYESSVVPGP KKAPMDSLFD YGTYRHHSSD NKRWRKKIIE KQPQSPKAPA PQPPPILKVF NRPILFDIVS R GSTADLDG ...String: MADSSEGPRA GPGEVAELPG DESGTPGGEA FPLSSLANLF EGEDGSLSPS PADASRPAGP GDGRPNLRMK FQGAFRKGVP NPIDLLEST LYESSVVPGP KKAPMDSLFD YGTYRHHSSD NKRWRKKIIE KQPQSPKAPA PQPPPILKVF NRPILFDIVS R GSTADLDG LLPFLLTHKK RLTDEEFREP STGKTCLPKA LLNLSNGRND TIPVLLDIAE RTGNMREFIN SPFRDIYYRG QT ALHIAIE RRCKHYVELL VAQGADVHAQ ARGRFFQPKD EGGYFYFGEL PLSLAACTNQ PHIVNYLTEN PHKKADMRRQ DSR GNTVLH ALVAIADNTR ENTKFVTKMY DLLLLKCARL FPDSNLEAVL NNDGLSPLMM AAKTGKIGIF QHIIRREVTD EDTR HLSRK FKDWAYGPVY SSLYDLSSLD TCGEEASVLE ILVYNSKIEN RHEMLAVEPI NELLRDKWRK FGAVSFYINV VSYLC AMVI FTLTAYYQPL EGTPPYPYRT TVDYLRLAGE VITLFTGVLF FFTNIKDLFM KKCPGVNSLF IDGSFQLLYF IYSVLV IVS AALYLAGIEA YLAVMVFALV LGWMNALYFT RGLKLTGTYS IMIQKILFKD LFRFLLVYLL FMIGYASALV SLLNPCA NM KVCNEDQTNC TVPTYPSCRD SETFSTFLLD LFKLTIGMGD LEMLSSTKYP VVFIILLVTY IILTFVLLLN MLIALMGE T VGQVSKESKH IWKLQWATTI LDIERSFPVF LRKAFRSGEM VTVGKSSDGT PDRRWCFRVD EVNWSHWNQN LGIINEDPG KNETYQYYGF SHTVGRLRRD RWSSVVPRVV ELNKNSNPDE VVVPLDSMGN PRCDGHQQGY PRKWRTDDAP LAAALEVLFQ GPSKGEELF TGVVPILVEL DGDVNGHKFS VRGEGEGDAT NGKLTLKFIC TTGKLPVPWP TLVTTLTYGV QCFSRYPDHM K RHDFFKSA MPEGYVQERT ISFKDDGTYK TRAEVKFEGD TLVNRIELKG IDFKEDGNIL GHKLEYNFNS HNVYITADKQ KN GIKANFK IRHNVEDGSV QLADHYQQNT PIGDGPVLLP DNHYLSTQSV LSKDPNEKRD HMVLLEFVTA AGITHGMDEW SHP QFEKGG GSGGGSGGSA WSHPQFEK UniProtKB: Transient receptor potential cation channel subfamily V member 4 |
-Macromolecule #2: 1-({(5S,7S)-3-[5-(2-hydroxypropan-2-yl)pyrazin-2-yl]-7-methyl-2-o...
| Macromolecule | Name: 1-({(5S,7S)-3-[5-(2-hydroxypropan-2-yl)pyrazin-2-yl]-7-methyl-2-oxo-1-oxa-3-azaspiro[4.5]decan-7-yl}methyl)-1H-benzimidazole-6-carbonitrile type: ligand / ID: 2 / Number of copies: 4 / Formula: XPW |
|---|---|
| Molecular weight | Theoretical: 460.528 Da |
| Chemical component information | 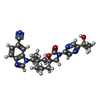 ChemComp-XPW: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)