[English] 日本語
 Yorodumi
Yorodumi- EMDB-36397: Cryo-EM structure of SV2A dimer in complex with BoNT/A2 Hc and le... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of SV2A dimer in complex with BoNT/A2 Hc and levetiracetam | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Synaptic vesicle / epilepsy / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationToxicity of botulinum toxin type F (botF) / Toxicity of botulinum toxin type D (botD) / Toxicity of botulinum toxin type E (botE) / Toxicity of botulinum toxin type A (botA) / presynaptic active zone / synaptic vesicle priming / transmembrane transporter activity / protein transmembrane transporter activity / neuromuscular junction / GABA-ergic synapse ...Toxicity of botulinum toxin type F (botF) / Toxicity of botulinum toxin type D (botD) / Toxicity of botulinum toxin type E (botE) / Toxicity of botulinum toxin type A (botA) / presynaptic active zone / synaptic vesicle priming / transmembrane transporter activity / protein transmembrane transporter activity / neuromuscular junction / GABA-ergic synapse / metalloendopeptidase activity / intracellular calcium ion homeostasis / synaptic vesicle / synaptic vesicle membrane / cell-cell junction / toxin activity / neuron projection / protein kinase binding / glutamatergic synapse / endoplasmic reticulum / proteolysis / extracellular region / zinc ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
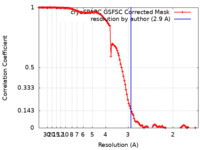
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Yamagata A | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis for antiepileptic drugs and botulinum neurotoxin recognition of SV2A. Authors: Atsushi Yamagata / Kaori Ito / Takehiro Suzuki / Naoshi Dohmae / Tohru Terada / Mikako Shirouzu /  Abstract: More than one percent of people have epilepsy worldwide. Levetiracetam (LEV) is a successful new-generation antiepileptic drug (AED), and its derivative, brivaracetam (BRV), shows improved efficacy. ...More than one percent of people have epilepsy worldwide. Levetiracetam (LEV) is a successful new-generation antiepileptic drug (AED), and its derivative, brivaracetam (BRV), shows improved efficacy. Synaptic vesicle glycoprotein 2a (SV2A), a putative membrane transporter in the synaptic vesicles (SVs), has been identified as a target of LEV and BRV. SV2A also serves as a receptor for botulinum neurotoxin (BoNT), which is the most toxic protein and has paradoxically emerged as a potent reagent for therapeutic and cosmetic applications. Nevertheless, no structural analysis on AEDs and BoNT recognition by full-length SV2A has been available. Here we describe the cryo-electron microscopy structures of the full-length SV2A in complex with the BoNT receptor-binding domain, BoNT/A2 H and either LEV or BRV. The large fourth luminal domain of SV2A binds to BoNT/A2 H through protein-protein and protein-glycan interactions. LEV and BRV occupy the putative substrate-binding site in an outward-open conformation. A propyl group in BRV creates additional contacts with SV2A, explaining its higher binding affinity than that of LEV, which was further supported by label-free spectral shift assay. Numerous LEV derivatives have been developed as AEDs and positron emission tomography (PET) tracers for neuroimaging. Our work provides a structural framework for AEDs and BoNT recognition of SV2A and a blueprint for the rational design of additional AEDs and PET tracers. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36397.map.gz emd_36397.map.gz | 168 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36397-v30.xml emd-36397-v30.xml emd-36397.xml emd-36397.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36397_fsc.xml emd_36397_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_36397.png emd_36397.png | 41.1 KB | ||
| Masks |  emd_36397_msk_1.map emd_36397_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-36397.cif.gz emd-36397.cif.gz | 6.7 KB | ||
| Others |  emd_36397_half_map_1.map.gz emd_36397_half_map_1.map.gz emd_36397_half_map_2.map.gz emd_36397_half_map_2.map.gz | 165.1 MB 165 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36397 http://ftp.pdbj.org/pub/emdb/structures/EMD-36397 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36397 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36397 | HTTPS FTP |
-Validation report
| Summary document |  emd_36397_validation.pdf.gz emd_36397_validation.pdf.gz | 894 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36397_full_validation.pdf.gz emd_36397_full_validation.pdf.gz | 893.6 KB | Display | |
| Data in XML |  emd_36397_validation.xml.gz emd_36397_validation.xml.gz | 20.2 KB | Display | |
| Data in CIF |  emd_36397_validation.cif.gz emd_36397_validation.cif.gz | 25.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36397 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36397 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36397 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36397 | HTTPS FTP |
-Related structure data
| Related structure data |  8jlhMC  8jlcC  8jleC  8jlfC  8jlgC  8jliC  8js8C  8js9C  8k77C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36397.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36397.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_36397_msk_1.map emd_36397_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_36397_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_36397_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SV2A dimer in complex with BoNT/A2 Hc domain and anti-epileptic d...
| Entire | Name: SV2A dimer in complex with BoNT/A2 Hc domain and anti-epileptic drug, levetiracetam |
|---|---|
| Components |
|
-Supramolecule #1: SV2A dimer in complex with BoNT/A2 Hc domain and anti-epileptic d...
| Supramolecule | Name: SV2A dimer in complex with BoNT/A2 Hc domain and anti-epileptic drug, levetiracetam type: complex / ID: 1 / Parent: 0 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Synaptic vesicle glycoprotein 2A
| Macromolecule | Name: Synaptic vesicle glycoprotein 2A / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 83.774008 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDYKDDDDKE EGFRDRAAFI RGAKDIAKEV KKHAAKKVVK GLDRVQDEYS RRSYSRFEEE DDDDDFPAPS DGYYRGEGTQ DEEEGGASS DATEGHDEDD EIYEGEYQGI PRAESGGKGE RMADGAPLAG VRGGLSDGEG PPGGRGEAQR RKEREELAQQ Y EAILRECG ...String: MDYKDDDDKE EGFRDRAAFI RGAKDIAKEV KKHAAKKVVK GLDRVQDEYS RRSYSRFEEE DDDDDFPAPS DGYYRGEGTQ DEEEGGASS DATEGHDEDD EIYEGEYQGI PRAESGGKGE RMADGAPLAG VRGGLSDGEG PPGGRGEAQR RKEREELAQQ Y EAILRECG HGRFQWTLYF VLGLALMADG VEVFVVGFVL PSAEKDMCLS DSNKGMLGLI VYLGMMVGAF LWGGLADRLG RR QCLLISL SVNSVFAFFS SFVQGYGTFL FCRLLSGVGI GGSIPIVFSY FSEFLAQEKR GEHLSWLCMF WMIGGVYAAA MAW AIIPHY GWSFQMGSAY QFHSWRVFVL VCAFPSVFAI GALTTQPESP RFFLENGKHD EAWMVLKQVH DTNMRAKGHP ERVF SVTHI KTIHQEDELI EIQSDTGTWY QRWGVRALSL GGQVWGNFLS CFGPEYRRIT LMMMGVWFTM SFSYYGLTVW FPDMI RHLQ AVDYASRTKV FPGERVEHVT FNFTLENQIH RGGQYFNDKF IGLRLKSVSF EDSLFEECYF EDVTSSNTFF RNCTFI NTV FYNTDLFEYK FVNSRLINST FLHNKEGCPL DVTGTGEGAY MVYFVSFLGT LAVLPGNIVS ALLMDKIGRL RMLAGSS VM SCVSCFFLSF GNSESAMIAL LCLFGGVSIA SWNALDVLTV ELYPSDKRTT AFGFLNALCK LAAVLGISIF TSFVGITK A APILFASAAL ALGSSLALKL PETRGQVLQ UniProtKB: Synaptic vesicle glycoprotein 2A |
-Macromolecule #2: Botulinum neurotoxin
| Macromolecule | Name: Botulinum neurotoxin / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 49.456191 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: KNIVNTSILS IVYKKDDLID LSRYGAKINI GDRVYYDSID KNQIKLINLE SSTIEVILKN AIVYNSMYEN FSTSFWIKIP KYFSKINLN NEYTIINCIE NNSGWKVSLN YGEIIWTLQD NKQNIQRVVF KYSQMVNISD YINRWIFVTI TNNRLTKSKI Y INGRLIDQ ...String: KNIVNTSILS IVYKKDDLID LSRYGAKINI GDRVYYDSID KNQIKLINLE SSTIEVILKN AIVYNSMYEN FSTSFWIKIP KYFSKINLN NEYTIINCIE NNSGWKVSLN YGEIIWTLQD NKQNIQRVVF KYSQMVNISD YINRWIFVTI TNNRLTKSKI Y INGRLIDQ KPISNLGNIH ASNKIMFKLD GCRDPRRYIM IKYFNLFDKE LNEKEIKDLY DSQSNSGILK DFWGNYLQYD KP YYMLNLF DPNKYVDVNN IGIRGYMYLK GPRGSVVTTN IYLNSTLYEG TKFIIKKYAS GNEDNIVRNN DRVYINVVVK NKE YRLATN ASQAGVEKIL SALEIPDVGN LSQVVVMKSK DDQGIRNKCK MNLQDNNGND IGFIGFHLYD NIAKLVASNW YNRQ VGKAS RTFGCSWEFI PVDDGWGESS L UniProtKB: Bontoxilysin-A |
-Macromolecule #4: (2S)-2-(2-oxidanylidenepyrrolidin-1-yl)butanamide
| Macromolecule | Name: (2S)-2-(2-oxidanylidenepyrrolidin-1-yl)butanamide / type: ligand / ID: 4 / Number of copies: 1 / Formula: UKX |
|---|---|
| Molecular weight | Theoretical: 170.209 Da |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 4 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Details: 20 mM Hepes (pH7.5), 150 mM NaCl, 0.001% LMNG, 0.0002% cholesterol hemisuccinate |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 297 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 6518 / Average electron dose: 50.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)