[English] 日本語
 Yorodumi
Yorodumi- EMDB-36934: Local map of SV2A LD4-BoNT/A2 Hc from SV2A-BoNT/A2 Hc-brivaraceta... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Local map of SV2A LD4-BoNT/A2 Hc from SV2A-BoNT/A2 Hc-brivaracetam complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Synapse / epilepsy / antiepileptic drug / botulinum neurotoxin / MEMBRANE PROTEIN | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
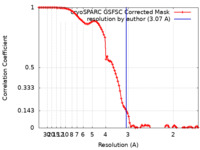
| Method | single particle reconstruction / cryo EM / Resolution: 3.07 Å | |||||||||
 Authors Authors | Yamagata A | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis for antiepileptic drugs and botulinum neurotoxin recognition of SV2A. Authors: Atsushi Yamagata / Kaori Ito / Takehiro Suzuki / Naoshi Dohmae / Tohru Terada / Mikako Shirouzu /  Abstract: More than one percent of people have epilepsy worldwide. Levetiracetam (LEV) is a successful new-generation antiepileptic drug (AED), and its derivative, brivaracetam (BRV), shows improved efficacy. ...More than one percent of people have epilepsy worldwide. Levetiracetam (LEV) is a successful new-generation antiepileptic drug (AED), and its derivative, brivaracetam (BRV), shows improved efficacy. Synaptic vesicle glycoprotein 2a (SV2A), a putative membrane transporter in the synaptic vesicles (SVs), has been identified as a target of LEV and BRV. SV2A also serves as a receptor for botulinum neurotoxin (BoNT), which is the most toxic protein and has paradoxically emerged as a potent reagent for therapeutic and cosmetic applications. Nevertheless, no structural analysis on AEDs and BoNT recognition by full-length SV2A has been available. Here we describe the cryo-electron microscopy structures of the full-length SV2A in complex with the BoNT receptor-binding domain, BoNT/A2 H and either LEV or BRV. The large fourth luminal domain of SV2A binds to BoNT/A2 H through protein-protein and protein-glycan interactions. LEV and BRV occupy the putative substrate-binding site in an outward-open conformation. A propyl group in BRV creates additional contacts with SV2A, explaining its higher binding affinity than that of LEV, which was further supported by label-free spectral shift assay. Numerous LEV derivatives have been developed as AEDs and positron emission tomography (PET) tracers for neuroimaging. Our work provides a structural framework for AEDs and BoNT recognition of SV2A and a blueprint for the rational design of additional AEDs and PET tracers. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36934.map.gz emd_36934.map.gz | 97 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36934-v30.xml emd-36934-v30.xml emd-36934.xml emd-36934.xml | 15.8 KB 15.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36934_fsc.xml emd_36934_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_36934.png emd_36934.png | 32.3 KB | ||
| Masks |  emd_36934_msk_1.map emd_36934_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-36934.cif.gz emd-36934.cif.gz | 5.5 KB | ||
| Others |  emd_36934_half_map_1.map.gz emd_36934_half_map_1.map.gz emd_36934_half_map_2.map.gz emd_36934_half_map_2.map.gz | 95.6 MB 95.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36934 http://ftp.pdbj.org/pub/emdb/structures/EMD-36934 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36934 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36934 | HTTPS FTP |
-Validation report
| Summary document |  emd_36934_validation.pdf.gz emd_36934_validation.pdf.gz | 869.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36934_full_validation.pdf.gz emd_36934_full_validation.pdf.gz | 868.9 KB | Display | |
| Data in XML |  emd_36934_validation.xml.gz emd_36934_validation.xml.gz | 17.3 KB | Display | |
| Data in CIF |  emd_36934_validation.cif.gz emd_36934_validation.cif.gz | 22 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36934 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36934 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36934 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36934 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36934.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36934.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_36934_msk_1.map emd_36934_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_36934_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_36934_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Synaptic vesicle glycroprotein 2A in complex with brivaracetam an...
| Entire | Name: Synaptic vesicle glycroprotein 2A in complex with brivaracetam and botulinum neurotoxin A2 Hc domain |
|---|---|
| Components |
|
-Supramolecule #1: Synaptic vesicle glycroprotein 2A in complex with brivaracetam an...
| Supramolecule | Name: Synaptic vesicle glycroprotein 2A in complex with brivaracetam and botulinum neurotoxin A2 Hc domain type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #2: Synaptic vesicle glycoprotein
| Supramolecule | Name: Synaptic vesicle glycoprotein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Botulinum neurotoxin A2
| Supramolecule | Name: Botulinum neurotoxin A2 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|
-Macromolecule #1: Synaptic vesicle glycoprotein
| Macromolecule | Name: Synaptic vesicle glycoprotein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MDYKDDDDKE EGFRDRAAFI RGAKDIAKEV KKHAAKKVVK GLDRVQDEYS RRSYSRFEEE DDDDDFPAP SDGYYRGEGT QDEEEGGASS DATEGHDEDD EIYEGEYQGI PRAESGGKGE R MADGAPLA GVRGGLSDGE GPPGGRGEAQ RRKEREELAQ QYEAILRECG ...String: MDYKDDDDKE EGFRDRAAFI RGAKDIAKEV KKHAAKKVVK GLDRVQDEYS RRSYSRFEEE DDDDDFPAP SDGYYRGEGT QDEEEGGASS DATEGHDEDD EIYEGEYQGI PRAESGGKGE R MADGAPLA GVRGGLSDGE GPPGGRGEAQ RRKEREELAQ QYEAILRECG HGRFQWTLYF VL GLALMAD GVEVFVVGFV LPSAEKDMCL SDSNKGMLGL IVYLGMMVGA FLWGGLADRL GRR QCLLIS LSVNSVFAFF SSFVQGYGTF LFCRLLSGVG IGGSIPIVFS YFSEFLAQEK RGEH LSWLC MFWMIGGVYA AAMAWAIIPH YGWSFQMGSA YQFHSWRVFV LVCAFPSVFA IGALT TQPE SPRFFLENGK HDEAWMVLKQ VHDTNMRAKG HPERVFSVTH IKTIHQEDEL IEIQSD TGT WYQRWGVRAL SLGGQVWGNF LSCFGPEYRR ITLMMMGVWF TMSFSYYGLT VWFPDMI RH LQAVDYASRT KVFPGERVEH VTFNFTLENQ IHRGGQYFND KFIGLRLKSV SFEDSLFE E CYFEDVTSSN TFFRNCTFIN TVFYNTDLFE YKFVNSRLIN STFLHNKEGC PLDVTGTGE GAYMVYFVSF LGTLAVLPGN IVSALLMDKI GRLRMLAGSS VMSCVSCFFL SFGNSESAMI ALLCLFGGV SIASWNALDV LTVELYPSDK RTTAFGFLNA LCKLAAVLGI SIFTSFVGIT K AAPILFAS AALALGSSLA LKLPETRGQV LQ |
-Macromolecule #2: Botulinum neurotoxin A2
| Macromolecule | Name: Botulinum neurotoxin A2 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: KNIVNTSILS IVYKKDDLID LSRYGAKINI GDRVYYDSID KNQIKLINLE SSTIEVILKN AIVYNSMYE NFSTSFWIKI PKYFSKINLN NEYTIINCIE NNSGWKVSLN YGEIIWTLQD N KQNIQRVV FKYSQMVNIS DYINRWIFVT ITNNRLTKSK IYINGRLIDQ ...String: KNIVNTSILS IVYKKDDLID LSRYGAKINI GDRVYYDSID KNQIKLINLE SSTIEVILKN AIVYNSMYE NFSTSFWIKI PKYFSKINLN NEYTIINCIE NNSGWKVSLN YGEIIWTLQD N KQNIQRVV FKYSQMVNIS DYINRWIFVT ITNNRLTKSK IYINGRLIDQ KPISNLGNIH AS NKIMFKL DGCRDPRRYI MIKYFNLFDK ELNEKEIKDL YDSQSNSGIL KDFWGNYLQY DKP YYMLNL FDPNKYVDVN NIGIRGYMYL KGPRGSVVTT NIYLNSTLYE GTKFIIKKYA SGNE DNIVR NNDRVYINVV VKNKEYRLAT NASQAGVEKI LSALEIPDVG NLSQVVVMKS KDDQG IRNK CKMNLQDNNG NDIGFIGFHL YDNIAKLVAS NWYNRQVGKA SRTFGCSWEF IPVDDG WGE SSL |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 53.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)