[English] 日本語
 Yorodumi
Yorodumi- EMDB-36180: Structure of the Human cytoplasmic Ribosome with human tRNA Tyr(G... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Human cytoplasmic Ribosome with human tRNA Tyr(GalQ34) and mRNA(UAU) (non-rotated state) | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | tRNA modifications / decoding / RIBOSOME | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationtranslation at presynapse / exit from mitosis / optic nerve development / response to insecticide / eukaryotic 80S initiation complex / negative regulation of protein neddylation / regulation of translation involved in cellular response to UV / axial mesoderm development / negative regulation of formation of translation preinitiation complex / regulation of G1 to G0 transition ...translation at presynapse / exit from mitosis / optic nerve development / response to insecticide / eukaryotic 80S initiation complex / negative regulation of protein neddylation / regulation of translation involved in cellular response to UV / axial mesoderm development / negative regulation of formation of translation preinitiation complex / regulation of G1 to G0 transition / retinal ganglion cell axon guidance / oxidized pyrimidine DNA binding / response to TNF agonist / negative regulation of endoplasmic reticulum unfolded protein response / positive regulation of base-excision repair / ribosomal protein import into nucleus / protein-DNA complex disassembly / positive regulation of respiratory burst involved in inflammatory response / positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator / positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage / positive regulation of gastrulation / 90S preribosome assembly / protein tyrosine kinase inhibitor activity / positive regulation of endodeoxyribonuclease activity / nucleolus organization / IRE1-RACK1-PP2A complex / positive regulation of Golgi to plasma membrane protein transport / TNFR1-mediated ceramide production / alpha-beta T cell differentiation / negative regulation of DNA repair / negative regulation of RNA splicing / GAIT complex / positive regulation of DNA damage response, signal transduction by p53 class mediator / TORC2 complex binding / G1 to G0 transition / supercoiled DNA binding / NF-kappaB complex / neural crest cell differentiation / oxidized purine DNA binding / cysteine-type endopeptidase activator activity involved in apoptotic process / middle ear morphogenesis / positive regulation of ubiquitin-protein transferase activity / negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide / negative regulation of bicellular tight junction assembly / regulation of establishment of cell polarity / ubiquitin-like protein conjugating enzyme binding / rRNA modification in the nucleus and cytosol / negative regulation of phagocytosis / erythrocyte homeostasis / Formation of the ternary complex, and subsequently, the 43S complex / cytoplasmic side of rough endoplasmic reticulum membrane / negative regulation of ubiquitin protein ligase activity / protein kinase A binding / laminin receptor activity / homeostatic process / ion channel inhibitor activity / Ribosomal scanning and start codon recognition / pigmentation / Translation initiation complex formation / positive regulation of mitochondrial depolarization / macrophage chemotaxis / lung morphogenesis / positive regulation of T cell receptor signaling pathway / fibroblast growth factor binding / negative regulation of Wnt signaling pathway / positive regulation of natural killer cell proliferation / male meiosis I / monocyte chemotaxis / TOR signaling / negative regulation of translational frameshifting / BH3 domain binding / positive regulation of activated T cell proliferation / Protein hydroxylation / SARS-CoV-1 modulates host translation machinery / iron-sulfur cluster binding / regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway / regulation of cell division / cellular response to ethanol / mTORC1-mediated signalling / Peptide chain elongation / Selenocysteine synthesis / Formation of a pool of free 40S subunits / positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator / cellular response to actinomycin D / endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / Eukaryotic Translation Termination / blastocyst development / positive regulation of GTPase activity / negative regulation of ubiquitin-dependent protein catabolic process / protein serine/threonine kinase inhibitor activity / SRP-dependent cotranslational protein targeting to membrane / Response of EIF2AK4 (GCN2) to amino acid deficiency / ubiquitin ligase inhibitor activity / Viral mRNA Translation / negative regulation of respiratory burst involved in inflammatory response / positive regulation of signal transduction by p53 class mediator / protein localization to nucleus / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / GTP hydrolysis and joining of the 60S ribosomal subunit / L13a-mediated translational silencing of Ceruloplasmin expression Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
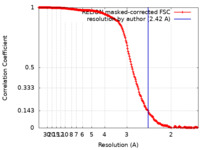
| Method | single particle reconstruction / cryo EM / Resolution: 2.42 Å | ||||||||||||
 Authors Authors | Ishiguro K / Yokoyama T / Shirouzu M / Suzuki T | ||||||||||||
| Funding support |  Japan, 3 items Japan, 3 items
| ||||||||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth. Authors: Xuewei Zhao / Ding Ma / Kensuke Ishiguro / Hironori Saito / Shinichiro Akichika / Ikuya Matsuzawa / Mari Mito / Toru Irie / Kota Ishibashi / Kimi Wakabayashi / Yuriko Sakaguchi / Takeshi ...Authors: Xuewei Zhao / Ding Ma / Kensuke Ishiguro / Hironori Saito / Shinichiro Akichika / Ikuya Matsuzawa / Mari Mito / Toru Irie / Kota Ishibashi / Kimi Wakabayashi / Yuriko Sakaguchi / Takeshi Yokoyama / Yuichiro Mishima / Mikako Shirouzu / Shintaro Iwasaki / Takeo Suzuki / Tsutomu Suzuki /  Abstract: Transfer RNA (tRNA) modifications are critical for protein synthesis. Queuosine (Q), a 7-deaza-guanosine derivative, is present in tRNA anticodons. In vertebrate tRNAs for Tyr and Asp, Q is further ...Transfer RNA (tRNA) modifications are critical for protein synthesis. Queuosine (Q), a 7-deaza-guanosine derivative, is present in tRNA anticodons. In vertebrate tRNAs for Tyr and Asp, Q is further glycosylated with galactose and mannose to generate galQ and manQ, respectively. However, biogenesis and physiological relevance of Q-glycosylation remain poorly understood. Here, we biochemically identified two RNA glycosylases, QTGAL and QTMAN, and successfully reconstituted Q-glycosylation of tRNAs using nucleotide diphosphate sugars. Ribosome profiling of knockout cells revealed that Q-glycosylation slowed down elongation at cognate codons, UAC and GAC (GAU), respectively. We also found that galactosylation of Q suppresses stop codon readthrough. Moreover, protein aggregates increased in cells lacking Q-glycosylation, indicating that Q-glycosylation contributes to proteostasis. Cryo-EM of human ribosome-tRNA complex revealed the molecular basis of codon recognition regulated by Q-glycosylations. Furthermore, zebrafish qtgal and qtman knockout lines displayed shortened body length, implying that Q-glycosylation is required for post-embryonic growth in vertebrates. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36180.map.gz emd_36180.map.gz | 516.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36180-v30.xml emd-36180-v30.xml emd-36180.xml emd-36180.xml | 104.3 KB 104.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36180_fsc.xml emd_36180_fsc.xml | 18.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_36180.png emd_36180.png | 78.1 KB | ||
| Filedesc metadata |  emd-36180.cif.gz emd-36180.cif.gz | 21.3 KB | ||
| Others |  emd_36180_additional_1.map.gz emd_36180_additional_1.map.gz emd_36180_half_map_1.map.gz emd_36180_half_map_1.map.gz emd_36180_half_map_2.map.gz emd_36180_half_map_2.map.gz | 455.4 MB 456.7 MB 456.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36180 http://ftp.pdbj.org/pub/emdb/structures/EMD-36180 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36180 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36180 | HTTPS FTP |
-Related structure data
| Related structure data |  8jdlMC  7y7cC  7y7dC  7y7eC  7y7fC  7y7gC  7y7hC  8jdjC  8jdkC  8jdmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36180.map.gz / Format: CCP4 / Size: 567.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36180.map.gz / Format: CCP4 / Size: 567.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
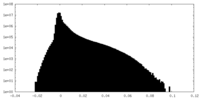
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: before postprocess
| File | emd_36180_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | before postprocess | ||||||||||||
| Projections & Slices |
| ||||||||||||
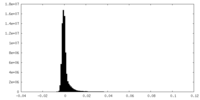
| Density Histograms |
-Half map: #2
| File | emd_36180_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
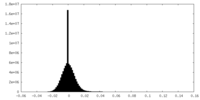
| Density Histograms |
-Half map: #1
| File | emd_36180_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
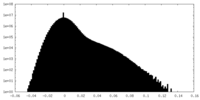
| Density Histograms |
- Sample components
Sample components
+Entire : The complex of Human 80S Ribosome with mRNA and P- site tRNA
+Supramolecule #1: The complex of Human 80S Ribosome with mRNA and P- site tRNA
+Macromolecule #1: mRNA
+Macromolecule #2: tRNA (Tyr)
+Macromolecule #3: 28S ribosomal RNA
+Macromolecule #4: 5S ribosomal RNA
+Macromolecule #5: 5.8S ribosomal RNA
+Macromolecule #48: 18S ribosomal RNA
+Macromolecule #6: 60S ribosomal protein L8
+Macromolecule #7: 60S ribosomal protein L3
+Macromolecule #8: 60S ribosomal protein L4
+Macromolecule #9: 60S ribosomal protein L5
+Macromolecule #10: 60S ribosomal protein L6
+Macromolecule #11: 60S ribosomal protein L7
+Macromolecule #12: 60S ribosomal protein L7a
+Macromolecule #13: 60S ribosomal protein L9
+Macromolecule #14: 60S ribosomal protein L10-like
+Macromolecule #15: 60S ribosomal protein L11
+Macromolecule #16: 60S ribosomal protein L13
+Macromolecule #17: 60S ribosomal protein L14
+Macromolecule #18: 60S ribosomal protein L15
+Macromolecule #19: 60S ribosomal protein L13a
+Macromolecule #20: 60S ribosomal protein L17
+Macromolecule #21: 60S ribosomal protein L18
+Macromolecule #22: 60S ribosomal protein L19
+Macromolecule #23: 60S ribosomal protein L18a
+Macromolecule #24: 60S ribosomal protein L21
+Macromolecule #25: 60S ribosomal protein L22
+Macromolecule #26: 60S ribosomal protein L23
+Macromolecule #27: 60S ribosomal protein L24
+Macromolecule #28: 60S ribosomal protein L23a
+Macromolecule #29: 60S ribosomal protein L26
+Macromolecule #30: 60S ribosomal protein L27
+Macromolecule #31: 60S ribosomal protein L27a
+Macromolecule #32: 60S ribosomal protein L29
+Macromolecule #33: 60S ribosomal protein L30
+Macromolecule #34: 60S ribosomal protein L31
+Macromolecule #35: 60S ribosomal protein L32
+Macromolecule #36: 60S ribosomal protein L35a
+Macromolecule #37: 60S ribosomal protein L34
+Macromolecule #38: 60S ribosomal protein L35
+Macromolecule #39: 60S ribosomal protein L36
+Macromolecule #40: 60S ribosomal protein L37
+Macromolecule #41: 60S ribosomal protein L38
+Macromolecule #42: 60S ribosomal protein L39
+Macromolecule #43: Ubiquitin-60S ribosomal protein L40
+Macromolecule #44: 60S ribosomal protein L41
+Macromolecule #45: 60S ribosomal protein L36a
+Macromolecule #46: 60S ribosomal protein L37a
+Macromolecule #47: 60S ribosomal protein L28
+Macromolecule #49: 40S ribosomal protein SA
+Macromolecule #50: 40S ribosomal protein S3a
+Macromolecule #51: 40S ribosomal protein S2
+Macromolecule #52: 40S ribosomal protein S3
+Macromolecule #53: 40S ribosomal protein S4, X isoform
+Macromolecule #54: 40S ribosomal protein S5
+Macromolecule #55: 40S ribosomal protein S6
+Macromolecule #56: 40S ribosomal protein S7
+Macromolecule #57: 40S ribosomal protein S8
+Macromolecule #58: 40S ribosomal protein S9
+Macromolecule #59: 40S ribosomal protein S10
+Macromolecule #60: 40S ribosomal protein S11
+Macromolecule #61: 40S ribosomal protein S13
+Macromolecule #62: 40S ribosomal protein S14
+Macromolecule #63: 40S ribosomal protein S15
+Macromolecule #64: 40S ribosomal protein S16
+Macromolecule #65: 40S ribosomal protein S17
+Macromolecule #66: 40S ribosomal protein S18
+Macromolecule #67: 40S ribosomal protein S19
+Macromolecule #68: 40S ribosomal protein S20
+Macromolecule #69: 40S ribosomal protein S21
+Macromolecule #70: 40S ribosomal protein S15a
+Macromolecule #71: 40S ribosomal protein S23
+Macromolecule #72: 40S ribosomal protein S24
+Macromolecule #73: 40S ribosomal protein S25
+Macromolecule #74: 40S ribosomal protein S26
+Macromolecule #75: 40S ribosomal protein S27
+Macromolecule #76: 40S ribosomal protein S28
+Macromolecule #77: 40S ribosomal protein S29
+Macromolecule #78: 40S ribosomal protein S30
+Macromolecule #79: Receptor of activated protein C kinase 1
+Macromolecule #80: MAGNESIUM ION
+Macromolecule #81: beta-D-galactopyranose
+Macromolecule #82: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
Details: The Buffer pH was adjusted to 7.4 using KOH. | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||
| Details | 24nM ribosomes were incubated with 500nM tRNAs and mRNA |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 2 / Number real images: 32000 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)