[English] 日本語
 Yorodumi
Yorodumi- EMDB-36125: Cryo-EM structure of Mycobacterium tuberculosis LpqY-SugABC in co... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Mycobacterium tuberculosis LpqY-SugABC in complex with trehalose | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ABC transporter / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationABC-type carbohydrate transporter activity / ATP-binding cassette (ABC) transporter complex, transmembrane substrate-binding subunit-containing / Translocases; Catalysing the translocation of carbohydrates and their derivatives; Linked to the hydrolysis of a nucleoside triphosphate / trehalose transmembrane transporter activity / trehalose transport / biological process involved in interaction with host / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / ATP-binding cassette (ABC) transporter complex / transmembrane transport / periplasmic space ...ABC-type carbohydrate transporter activity / ATP-binding cassette (ABC) transporter complex, transmembrane substrate-binding subunit-containing / Translocases; Catalysing the translocation of carbohydrates and their derivatives; Linked to the hydrolysis of a nucleoside triphosphate / trehalose transmembrane transporter activity / trehalose transport / biological process involved in interaction with host / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / ATP-binding cassette (ABC) transporter complex / transmembrane transport / periplasmic space / protein homodimerization activity / ATP hydrolysis activity / ATP binding / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.02 Å | |||||||||
 Authors Authors | Zhang B / Liang J / Rao Z | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Molecular recognition of trehalose and trehalose analogues by LpqY-SugABC. Authors: Jingxi Liang / Fengjiang Liu / Peng Xu / Wei Shangguan / Tianyu Hu / Shule Wang / Xiaolin Yang / Zhiqi Xiong / Xiuna Yang / Luke W Guddat / Biao Yu / Zihe Rao / Bing Zhang /   Abstract: Trehalose plays a crucial role in the survival and virulence of the deadly human pathogen (). The type I ATP-binding cassette (ABC) transporter LpqY-SugABC is the sole pathway for trehalose to enter ...Trehalose plays a crucial role in the survival and virulence of the deadly human pathogen (). The type I ATP-binding cassette (ABC) transporter LpqY-SugABC is the sole pathway for trehalose to enter . The substrate-binding protein, LpqY, which forms a stable complex with the translocator SugABC, recognizes and captures trehalose and its analogues in the periplasmic space, but the precise molecular mechanism for this process is still not well understood. This study reports a 3.02-Å cryoelectron microscopy structure of trehalose-bound LpqY-SugABC in the pretranslocation state, a crystal structure of LpqY in a closed form with trehalose bound and five crystal structures of LpqY in complex with different trehalose analogues. These structures, accompanied by substrate-stimulated ATPase activity data, reveal how LpqY recognizes and binds trehalose and its analogues, and highlight the flexibility in the substrate binding pocket of LpqY. These data provide critical insights into the design of trehalose analogues that could serve as potential molecular probe tools or as anti-TB drugs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36125.map.gz emd_36125.map.gz | 107.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36125-v30.xml emd-36125-v30.xml emd-36125.xml emd-36125.xml | 21 KB 21 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36125.png emd_36125.png | 35.6 KB | ||
| Filedesc metadata |  emd-36125.cif.gz emd-36125.cif.gz | 6.9 KB | ||
| Others |  emd_36125_half_map_1.map.gz emd_36125_half_map_1.map.gz emd_36125_half_map_2.map.gz emd_36125_half_map_2.map.gz | 200.2 MB 200.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36125 http://ftp.pdbj.org/pub/emdb/structures/EMD-36125 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36125 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36125 | HTTPS FTP |
-Related structure data
| Related structure data |  8ja7MC  8ja8C  8ja9C  8jaaC  8jabC  8jacC  8jadC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36125.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36125.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
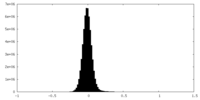
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_36125_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
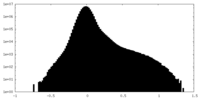
| Density Histograms |
-Half map: #1
| File | emd_36125_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
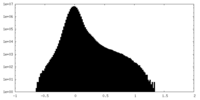
| Density Histograms |
- Sample components
Sample components
-Entire : LpqY-SugABC
| Entire | Name: LpqY-SugABC |
|---|---|
| Components |
|
-Supramolecule #1: LpqY-SugABC
| Supramolecule | Name: LpqY-SugABC / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) |
-Macromolecule #1: Trehalose transport system permease protein SugA
| Macromolecule | Name: Trehalose transport system permease protein SugA / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) |
| Molecular weight | Theoretical: 33.041809 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Sequence | String: VTSVEQRTAT AVFSRTGSRM AERRLAFMLV APAAMLMVAV TAYPIGYALW LSLQRNNLAT PNDTAFIGLG NYHTILIDRY WWTALAVTL AITAVSVTIE FVLGLALALV MHRTLIGKGL VRTAVLIPYG IVTVVASYSW YYAWTPGTGY LANLLPYDSA P LTQQIPSL ...String: VTSVEQRTAT AVFSRTGSRM AERRLAFMLV APAAMLMVAV TAYPIGYALW LSLQRNNLAT PNDTAFIGLG NYHTILIDRY WWTALAVTL AITAVSVTIE FVLGLALALV MHRTLIGKGL VRTAVLIPYG IVTVVASYSW YYAWTPGTGY LANLLPYDSA P LTQQIPSL GIVVIAEVWK TTPFMSLLLL AGLALVPEDL LRAAQVDGAS AWRRLTKVIL PMIKPAIVVA LLFRTLDAFR IF DNIYVLT GGSNNTGSVS ILGYDNLFKG FNVGLGSAIS VLIFGCVAVI AFIFIKLFGA AAPGGEPSGR UniProtKB: Trehalose transport system permease protein SugA |
-Macromolecule #2: Trehalose transport system permease protein SugB
| Macromolecule | Name: Trehalose transport system permease protein SugB / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) |
| Molecular weight | Theoretical: 29.145537 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Sequence | String: VGARRATYWA VLDTLVVGYA LLPVLWIFSL SLKPTSTVKD GKLIPSTVTF DNYRGIFRGD LFSSALINSI GIGLITTVIA VVLGAMAAY AVARLEFPGK RLLIGAALLI TMFPSISLVT PLFNIERAIG LFDTWPGLIL PYITFALPLA IYTLSAFFRE I PWDLEKAA ...String: VGARRATYWA VLDTLVVGYA LLPVLWIFSL SLKPTSTVKD GKLIPSTVTF DNYRGIFRGD LFSSALINSI GIGLITTVIA VVLGAMAAY AVARLEFPGK RLLIGAALLI TMFPSISLVT PLFNIERAIG LFDTWPGLIL PYITFALPLA IYTLSAFFRE I PWDLEKAA KMDGATPGQA FRKVIVPLAA PGLVTAAILV FIFAWNDLLL ALSLTATKAA ITAPVAIANF TGSSQFEEPT GS IAAGAIV ITIPIIVFVL IFQRRIVAGL TSGAVKG UniProtKB: Trehalose transport system permease protein SugB |
-Macromolecule #3: Trehalose-binding lipoprotein LpqY
| Macromolecule | Name: Trehalose-binding lipoprotein LpqY / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) |
| Molecular weight | Theoretical: 49.803074 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Sequence | String: VVMSRGRIPR LGAAVLVALT TAAAACGADS QGLVVSFYTP ATDGATFTAI AQRCNQQFGG RFTIAQVSLP RSPNEQRLQL ARRLTGNDR TLDVMALDVV WTAEFAEAGW ALPLSDDPAG LAENDAVADT LPGPLATAGW NHKLYAAPVT TNTQLLWYRP D LVNSPPTD ...String: VVMSRGRIPR LGAAVLVALT TAAAACGADS QGLVVSFYTP ATDGATFTAI AQRCNQQFGG RFTIAQVSLP RSPNEQRLQL ARRLTGNDR TLDVMALDVV WTAEFAEAGW ALPLSDDPAG LAENDAVADT LPGPLATAGW NHKLYAAPVT TNTQLLWYRP D LVNSPPTD WNAMIAEAAR LHAAGEPSWI AVQANQGEGL VVWFNTLLVS AGGSVLSEDG RHVTLTDTPA HRAATVSALQ IL KSVATTP GADPSITRTE EGSARLAFEQ GKAALEVNWP FVFASMLENA VKGGVPFLPL NRIPQLAGSI NDIGTFTPSD EQF RIAYDA SQQVFGFAPY PAVAPGQPAK VTIGGLNLAV AKTTRHRAEA FEAVRCLRDQ HNQRYVSLEG GLPAVRASLY SDPQ FQAKY PMHAIIRQQL TDAAVRPATP VYQALSIRLA AVLSPITEID PESTADELAA QAQKAIDGMG LLP UniProtKB: Trehalose-binding lipoprotein LpqY |
-Macromolecule #4: Trehalose import ATP-binding protein SugC
| Macromolecule | Name: Trehalose import ATP-binding protein SugC / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO EC number: Translocases; Catalysing the translocation of carbohydrates and their derivatives; Linked to the hydrolysis of a nucleoside triphosphate |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) |
| Molecular weight | Theoretical: 42.964078 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Sequence | String: MAEIVLDHVN KSYPDGHTAV RDLNLTIADG EFLILVGPSG CGKTTTLNMI AGLEDISSGE LRIAGERVNE KAPKDRDIAM VFQSYALYP HMTVRQNIAF PLTLAKMRKA DIAQKVSETA KILDLTNLLD RKPSQLSGGQ RQRVAMGRAI VRHPKAFLMD E PLSNLDAK ...String: MAEIVLDHVN KSYPDGHTAV RDLNLTIADG EFLILVGPSG CGKTTTLNMI AGLEDISSGE LRIAGERVNE KAPKDRDIAM VFQSYALYP HMTVRQNIAF PLTLAKMRKA DIAQKVSETA KILDLTNLLD RKPSQLSGGQ RQRVAMGRAI VRHPKAFLMD E PLSNLDAK LRVQMRGEIA QLQRRLGTTT VYVTHDQTEA MTLGDRVVVM YGGIAQQIGT PEELYERPAN LFVAGFIGSP AM NFFPARL TAIGLTLPFG EVTLAPEVQG VIAAHPKPEN VIVGVRPEHI QDAALIDAYQ RIRALTFQVK VNLVESLGAD KYL YFTTES PAVHSVQLDE LAEVEGESAL HENQFVARVP AESKVAIGQS VELAFDTARL AVFDADSGAN LTIPHRA UniProtKB: Trehalose import ATP-binding protein SugC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)