[English] 日本語
 Yorodumi
Yorodumi- EMDB-3611: Full-length dodecameric S. typhimurium Wzz complex with associate... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3611 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Full-length dodecameric S. typhimurium Wzz complex with associated dodecyl maltoside micelle. | |||||||||
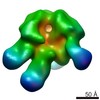
 Map data Map data | Wzz dodecamer with dodecyl maltoside micelle | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Wzz Polysaccharide Chain Length membrane protein Cryo-electron microscopy Single particle analysis / Membrane protein | |||||||||
| Function / homology | Polysaccharide chain length determinant N-terminal domain / Chain length determinant protein / : / lipopolysaccharide biosynthetic process / plasma membrane / Chain length determinant protein Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 9.0 Å | |||||||||
 Authors Authors | Ford RC / Kargas V | |||||||||
 Citation Citation |  Journal: Structure / Year: 2017 Journal: Structure / Year: 2017Title: Full-length, Oligomeric Structure of Wzz Determined by Cryoelectron Microscopy Reveals Insights into Membrane-Bound States. Authors: Richard F Collins / Vasileios Kargas / Brad R Clarke / C Alistair Siebert / Daniel K Clare / Peter J Bond / Chris Whitfield / Robert C Ford /    Abstract: Wzz is an integral inner membrane protein involved in regulating the length of lipopolysaccharide O-antigen glycans and essential for the virulence of many Gram-negative pathogens. In all Wzz ...Wzz is an integral inner membrane protein involved in regulating the length of lipopolysaccharide O-antigen glycans and essential for the virulence of many Gram-negative pathogens. In all Wzz homologs, the large periplasmic domain is proposed to be anchored by two transmembrane helices, but no information is available for the transmembrane and cytosolic domains. Here we have studied purified oligomeric Wzz complexes using cryoelectron microscopy and resolved the transmembrane regions within a semi-continuous detergent micelle. The transmembrane helices of each monomer display a right-handed super-helical twist, and do not interact with the neighboring transmembrane domains. Modeling, flexible fitting and multiscale simulation approaches were used to study the full-length complex and to provide explanations for the influence of the lipid bilayer on its oligomeric status. Based on structural and in silico observations, we propose a new mechanism for O-antigen chain-length regulation that invokes synergy of Wzz and its polymerase partner, Wzy. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3611.map.gz emd_3611.map.gz | 8.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3611-v30.xml emd-3611-v30.xml emd-3611.xml emd-3611.xml | 11.1 KB 11.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3611.png emd_3611.png | 216 KB | ||
| Filedesc metadata |  emd-3611.cif.gz emd-3611.cif.gz | 5.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3611 http://ftp.pdbj.org/pub/emdb/structures/EMD-3611 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3611 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3611 | HTTPS FTP |
-Related structure data
| Related structure data |  5nbzMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3611.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3611.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Wzz dodecamer with dodecyl maltoside micelle | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.45 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Dodecameric complex
| Entire | Name: Dodecameric complex |
|---|---|
| Components |
|
-Supramolecule #1: Dodecameric complex
| Supramolecule | Name: Dodecameric complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 360 KDa |
-Macromolecule #1: WzzB
| Macromolecule | Name: WzzB / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.760129 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: KWTSTAIITQ PDVGQIAGYN NAMNVIYGQA APKVSDLQET LIGRFSSAFS ALAETLDNQE EPEKLTIEPS VKNQQLPLTV SYVGQTAEG AQMKLAQYIQ QVDDKVNQEL ERDLKDNIAL GRKNLQDSLR TQEVVAQEQK DLRIRQIEEA LRYADEAKIT Q PQIQQTQD ...String: KWTSTAIITQ PDVGQIAGYN NAMNVIYGQA APKVSDLQET LIGRFSSAFS ALAETLDNQE EPEKLTIEPS VKNQQLPLTV SYVGQTAEG AQMKLAQYIQ QVDDKVNQEL ERDLKDNIAL GRKNLQDSLR TQEVVAQEQK DLRIRQIEEA LRYADEAKIT Q PQIQQTQD VTQDTMFLLG SDALKSMIQN EATRPLAFSP AYYQTKQTLL DIKNLKVTAD TVHVYRYVMK PTLPVRRDS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 20mM Tris pH 7.5, 150mM NaCl, 0.025% dodecyl maltoside. |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK III / Details: 1 x 4sec blot. |
| Details | Dodecamers with C12 symmetry with a small fraction of head-to-head D12 complexes. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 9.0 Å / Resolution method: OTHER / Details: ResMap / Number images used: 22000 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















