[English] 日本語
 Yorodumi
Yorodumi- EMDB-1245: Structural analysis of the anaphase-promoting complex reveals mul... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1245 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural analysis of the anaphase-promoting complex reveals multiple active sites and insights into polyubiquitylation. | |||||||||
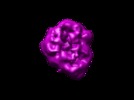
 Map data Map data | Structure of Saccharomyces cerevisiae Anaphase-Promoting Complex/Cyclosome (APC). Due to heterogeneity, we obtained 2 structures (A and B) from our dataset. This is "structure B" while EMD-1174 is "structure A". | |||||||||
 Sample Sample |
| |||||||||
| Function / homology | anaphase-promoting complex Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 20.0 Å | |||||||||
 Authors Authors | Passmore LA / Booth CR / Venien-Bryan C / Ludtke SJ / Fioretto C / Johnson LN / Chiu W / Barford D | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2005 Journal: Mol Cell / Year: 2005Title: Structural analysis of the anaphase-promoting complex reveals multiple active sites and insights into polyubiquitylation. Authors: Lori A Passmore / Christopher R Booth / Catherine Vénien-Bryan / Steven J Ludtke / Céline Fioretto / Louise N Johnson / Wah Chiu / David Barford /  Abstract: The anaphase-promoting complex/cyclosome (APC/C) is an E3 ubiquitin ligase composed of approximately 13 distinct subunits required for progression through meiosis, mitosis, and the G1 phase of the ...The anaphase-promoting complex/cyclosome (APC/C) is an E3 ubiquitin ligase composed of approximately 13 distinct subunits required for progression through meiosis, mitosis, and the G1 phase of the cell cycle. Despite its central role in these processes, information concerning its composition and structure is limited. Here, we determined the structure of yeast APC/C by cryo-electron microscopy (cryo-EM). Docking of tetratricopeptide repeat (TPR)-containing subunits indicates that they likely form a scaffold-like outer shell, mediating assembly of the complex and providing potential binding sites for regulators and substrates. Quantitative determination of subunit stoichiometry indicates multiple copies of specific subunits, consistent with a total APC/C mass of approximately 1.7 MDa. Moreover, yeast APC/C forms both monomeric and dimeric species. Dimeric APC/C is a more active E3 ligase than the monomer, with greatly enhanced processivity. Our data suggest that multimerisation and/or the presence of multiple active sites facilitates the APC/C's ability to elongate polyubiquitin chains. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1245.map.gz emd_1245.map.gz | 6.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1245-v30.xml emd-1245-v30.xml emd-1245.xml emd-1245.xml | 11 KB 11 KB | Display Display |  EMDB header EMDB header |
| Images |  1245.gif 1245.gif | 6.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1245 http://ftp.pdbj.org/pub/emdb/structures/EMD-1245 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1245 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1245 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1245.map.gz / Format: CCP4 / Size: 11.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1245.map.gz / Format: CCP4 / Size: 11.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of Saccharomyces cerevisiae Anaphase-Promoting Complex/Cyclosome (APC). Due to heterogeneity, we obtained 2 structures (A and B) from our dataset. This is "structure B" while EMD-1174 is "structure A". | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.168 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Anaphase-Promoting Complex
| Entire | Name: Anaphase-Promoting Complex |
|---|---|
| Components |
|
-Supramolecule #1000: Anaphase-Promoting Complex
| Supramolecule | Name: Anaphase-Promoting Complex / type: sample / ID: 1000 Details: The APC was freshly purified before grid preparation. Oligomeric state: monomer / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 1.7 MDa / Theoretical: 1.7 MDa |
-Macromolecule #1: Anaphase-Promoting Complex, Cyclosome
| Macromolecule | Name: Anaphase-Promoting Complex, Cyclosome / type: protein_or_peptide / ID: 1 / Name.synonym: APC Details: Endogenous APC was purified using a TAP tag present at the C-terminus of the Cdc16 subunit. The APC has 13 different protein subunits. There are multiple copies of specific APC subunits, ...Details: Endogenous APC was purified using a TAP tag present at the C-terminus of the Cdc16 subunit. The APC has 13 different protein subunits. There are multiple copies of specific APC subunits, consistent with a total mass of 1.7 MDa. Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 1.7 MDa / Theoretical: 1.7 MDa |
| Recombinant expression | Organism:  |
| Sequence | GO: anaphase-promoting complex |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.125 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 20 mM HEPES, 150 mM NaCl, 2 mM EGTA, 3 mM DTT, 1 mM Magnesium acetate, 0.015% (w/v) n-Dodecyl B-D-maltoside (DDM) |
| Grid | Details: Quantifoil R2/2, 200 mesh with an additional layer of freshly floated carbon |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Instrument: OTHER / Details: Vitrification instrument: Vitrobot Method: 4 ul sample was applied to the grid then blotted two times one second on both sides before plunging into liquid ethane. |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2010F |
|---|---|
| Temperature | Average: 92 K |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Digitization - Sampling interval: 15 µm / Number real images: 650 / Average electron dose: 15 e/Å2 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 69200 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 1 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.8 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Side entry liquid-nitrogen cooled cryo specimen holder Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| CTF correction | Details: Complete phase flipping and amplitude weighting with wiener filtration using the 1D structure factor |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 20.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: eman Details: Due to sample heterogeneity, the data could be separated into two main groups using a multirefinement procedure. Structure A (7500 particles) is in EMD-1174. This is structure B (6800 particles). Number images used: 19384 |
-Atomic model buiding 1
| Software | Name: Foldhunter, DockEM |
|---|---|
| Details | Protocol: rigid body. 15 or 18 consecutive TPR motifs modelled from PP5 (1A17) were docked as rigid structures into the APC density map using Foldhunter or DockEM. Both programs produced similar results. |
| Refinement | Protocol: RIGID BODY FIT / Target criteria: Cross-correlation |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















