+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
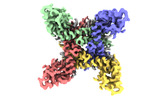
| Title | ion channel | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ion channel / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationvoltage-gated sodium channel complex / voltage-gated sodium channel activity / bioluminescence / generation of precursor metabolites and energy / calcium ion binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Chen HW / Jiang D | |||||||||
| Funding support |  Brazil, 1 items Brazil, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural mechanism of voltage-gated sodium channel slow inactivation. Authors: Huiwen Chen / Zhanyi Xia / Jie Dong / Bo Huang / Jiangtao Zhang / Feng Zhou / Rui Yan / Yiqiang Shi / Jianke Gong / Juquan Jiang / Zhuo Huang / Daohua Jiang /  Abstract: Voltage-gated sodium (Na) channels mediate a plethora of electrical activities. Na channels govern cellular excitability in response to depolarizing stimuli. Inactivation is an intrinsic property of ...Voltage-gated sodium (Na) channels mediate a plethora of electrical activities. Na channels govern cellular excitability in response to depolarizing stimuli. Inactivation is an intrinsic property of Na channels that regulates cellular excitability by controlling the channel availability. The fast inactivation, mediated by the Ile-Phe-Met (IFM) motif and the N-terminal helix (N-helix), has been well-characterized. However, the molecular mechanism underlying Na channel slow inactivation remains elusive. Here, we demonstrate that the removal of the N-helix of NaEh (NaEh) results in a slow-inactivated channel, and present cryo-EM structure of NaEh in a potential slow-inactivated state. The structure features a closed activation gate and a dilated selectivity filter (SF), indicating that the upper SF and the inner gate could serve as a gate for slow inactivation. In comparison to the NaEh structure, NaEh undergoes marked conformational shifts on the intracellular side. Together, our results provide important mechanistic insights into Na channel slow inactivation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36042.map.gz emd_36042.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36042-v30.xml emd-36042-v30.xml emd-36042.xml emd-36042.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36042.png emd_36042.png | 70.1 KB | ||
| Filedesc metadata |  emd-36042.cif.gz emd-36042.cif.gz | 6.6 KB | ||
| Others |  emd_36042_half_map_1.map.gz emd_36042_half_map_1.map.gz emd_36042_half_map_2.map.gz emd_36042_half_map_2.map.gz | 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36042 http://ftp.pdbj.org/pub/emdb/structures/EMD-36042 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36042 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36042 | HTTPS FTP |
-Validation report
| Summary document |  emd_36042_validation.pdf.gz emd_36042_validation.pdf.gz | 935.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36042_full_validation.pdf.gz emd_36042_full_validation.pdf.gz | 935.4 KB | Display | |
| Data in XML |  emd_36042_validation.xml.gz emd_36042_validation.xml.gz | 12.3 KB | Display | |
| Data in CIF |  emd_36042_validation.cif.gz emd_36042_validation.cif.gz | 14.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36042 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36042 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36042 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36042 | HTTPS FTP |
-Related structure data
| Related structure data |  8j7mMC  8j7fC  8j7hC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36042.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36042.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
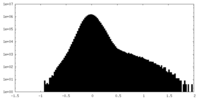
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_36042_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
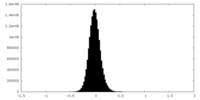
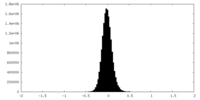
| Density Histograms |
-Half map: #2
| File | emd_36042_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
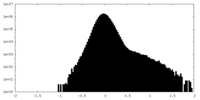
| Density Histograms |
- Sample components
Sample components
-Entire : ion channel
| Entire | Name: ion channel |
|---|---|
| Components |
|
-Supramolecule #1: ion channel
| Supramolecule | Name: ion channel / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: ion channel,Voltage dependent ion channel,Green fluorescent prote...
| Macromolecule | Name: ion channel,Voltage dependent ion channel,Green fluorescent protein (Fragment),Voltage dependent ion channel,Green fluorescent protein (Fragment),Voltage dependent ion channel,Green fluorescent protein (Fragment) type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 90.262719 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEAAAAHKAQ HRTAENSMDS LEDSTHETDA GERAQAGSTK LAWTDVVAPP PRKVVFWLPH QRKVFDFYAS QGVQYFTAFL IVSNFIFNC AEKEWDPYTD QLYQGLWRWG EFAFNTMFLI ELLINFYGIA FCFWRYNWAW NTFDLVVVAI GTLTMAEAIG G NFMPPSMA ...String: MEAAAAHKAQ HRTAENSMDS LEDSTHETDA GERAQAGSTK LAWTDVVAPP PRKVVFWLPH QRKVFDFYAS QGVQYFTAFL IVSNFIFNC AEKEWDPYTD QLYQGLWRWG EFAFNTMFLI ELLINFYGIA FCFWRYNWAW NTFDLVVVAI GTLTMAEAIG G NFMPPSMA LIRNLRAFRI FRLFKRIKSL NKIIVSLGKA IPGVANAFVI MVIIMCIYAI LGVEFYHMTG SDGTYVTYND NV KRGLCTG DEVELGQCSL NQTVSSETAR GYTYGEEYYG TFFRALYTLF QVLTGESWSE AVARPAVFES HYDSFGPVLF YVS FIIICQ IVLINVVVAV LLDKMVEEDD SEDPEKQTVA EKLSEMLSQE HAQLREIFRT WDEDNSGTIS IKEWRKAVKS MGYR GPIDV LDQIFASMDK DHSGELDYAE IDRMLSPTAA RERRSSTHAN PKRSVKEEVV AMRAEFTDHV ARLETQIAAL VLELQ LQRK PCGAEAPAPA HSRLAHDSDG APTEPPPPAA PDHHHLEDDE DTTQRVAAAL EVLFQGPSKG EELFTGVVPI LVELDG DVN GHKFSVRGEG EGDATNGKLT LKFICTTGKL PVPWPTLVTT LTYGVQCFSR YPDHMKRHDF FKSAMPEGYV QERTISF KD DGTYKTRAEV KFEGDTLVNR IELKGIDFKE DGNILGHKLE YNFNSHNVYI TADKQKNGIK ANFKIRHNVE DGSVQLAD H YQQNTPIGDG PVLLPDNHYL STQSVLSKDP NEKRDHMVLL EFVTAAGITH GMDEWSHPQF EKGGGSGGGS GGSAWSHPQ FEK UniProtKB: Voltage dependent ion channel, Green fluorescent protein |
-Macromolecule #2: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(tri...
| Macromolecule | Name: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate type: ligand / ID: 2 / Number of copies: 4 / Formula: POV |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-POV: |
-Macromolecule #3: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 3 / Number of copies: 10 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)