+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human Nav1.7 in complex with Hardwickii acid | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Inhibitor complex. / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcorticospinal neuron axon guidance / positive regulation of voltage-gated sodium channel activity / action potential propagation / detection of mechanical stimulus involved in sensory perception / regulation of atrial cardiac muscle cell membrane depolarization / membrane depolarization during Purkinje myocyte cell action potential / cardiac conduction / membrane depolarization during cardiac muscle cell action potential / membrane depolarization during action potential / positive regulation of sodium ion transport ...corticospinal neuron axon guidance / positive regulation of voltage-gated sodium channel activity / action potential propagation / detection of mechanical stimulus involved in sensory perception / regulation of atrial cardiac muscle cell membrane depolarization / membrane depolarization during Purkinje myocyte cell action potential / cardiac conduction / membrane depolarization during cardiac muscle cell action potential / membrane depolarization during action potential / positive regulation of sodium ion transport / regulation of sodium ion transmembrane transport / regulation of ventricular cardiac muscle cell membrane repolarization / cardiac muscle cell action potential involved in contraction / node of Ranvier / voltage-gated sodium channel complex / sodium channel inhibitor activity / locomotion / neuronal action potential propagation / Interaction between L1 and Ankyrins / voltage-gated sodium channel activity / detection of temperature stimulus involved in sensory perception of pain / Phase 0 - rapid depolarisation / regulation of heart rate by cardiac conduction / behavioral response to pain / intercalated disc / membrane depolarization / sodium channel regulator activity / neuronal action potential / cardiac muscle contraction / axon terminus / sensory perception of pain / T-tubule / axon guidance / sodium ion transmembrane transport / post-embryonic development / positive regulation of neuron projection development / circadian rhythm / response to toxic substance / Sensory perception of sweet, bitter, and umami (glutamate) taste / perikaryon / transmembrane transporter binding / cell adhesion / inflammatory response / axon / extracellular region / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Wu QR / Yan N | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural mapping of Na1.7 antagonists. Authors: Qiurong Wu / Jian Huang / Xiao Fan / Kan Wang / Xueqin Jin / Gaoxingyu Huang / Jiaao Li / Xiaojing Pan / Nieng Yan /   Abstract: Voltage-gated sodium (Na) channels are targeted by a number of widely used and investigational drugs for the treatment of epilepsy, arrhythmia, pain, and other disorders. Despite recent advances in ...Voltage-gated sodium (Na) channels are targeted by a number of widely used and investigational drugs for the treatment of epilepsy, arrhythmia, pain, and other disorders. Despite recent advances in structural elucidation of Na channels, the binding mode of most Na-targeting drugs remains unknown. Here we report high-resolution cryo-EM structures of human Na1.7 treated with drugs and lead compounds with representative chemical backbones at resolutions of 2.6-3.2 Å. A binding site beneath the intracellular gate (site BIG) accommodates carbamazepine, bupivacaine, and lacosamide. Unexpectedly, a second molecule of lacosamide plugs into the selectivity filter from the central cavity. Fenestrations are popular sites for various state-dependent drugs. We show that vinpocetine, a synthetic derivative of a vinca alkaloid, and hardwickiic acid, a natural product with antinociceptive effect, bind to the III-IV fenestration, while vixotrigine, an analgesic candidate, penetrates the IV-I fenestration of the pore domain. Our results permit building a 3D structural map for known drug-binding sites on Na channels summarized from the present and previous structures. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35975.map.gz emd_35975.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35975-v30.xml emd-35975-v30.xml emd-35975.xml emd-35975.xml | 18.5 KB 18.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35975.png emd_35975.png | 67 KB | ||
| Filedesc metadata |  emd-35975.cif.gz emd-35975.cif.gz | 7.4 KB | ||
| Others |  emd_35975_half_map_1.map.gz emd_35975_half_map_1.map.gz emd_35975_half_map_2.map.gz emd_35975_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35975 http://ftp.pdbj.org/pub/emdb/structures/EMD-35975 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35975 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35975 | HTTPS FTP |
-Validation report
| Summary document |  emd_35975_validation.pdf.gz emd_35975_validation.pdf.gz | 827.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35975_full_validation.pdf.gz emd_35975_full_validation.pdf.gz | 827.1 KB | Display | |
| Data in XML |  emd_35975_validation.xml.gz emd_35975_validation.xml.gz | 12.4 KB | Display | |
| Data in CIF |  emd_35975_validation.cif.gz emd_35975_validation.cif.gz | 14.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35975 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35975 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35975 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35975 | HTTPS FTP |
-Related structure data
| Related structure data |  8j4fMC  8i5bC  8i5gC  8i5xC  8i5yC  8s9bC  8s9cC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35975.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35975.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0979 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35975_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
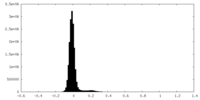
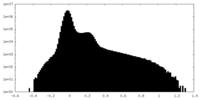
| Density Histograms |
-Half map: #1
| File | emd_35975_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
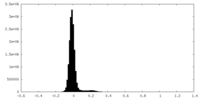
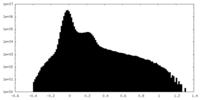
| Density Histograms |
- Sample components
Sample components
-Entire : human Nav1.7 in complex with hardwickii acid
| Entire | Name: human Nav1.7 in complex with hardwickii acid |
|---|---|
| Components |
|
-Supramolecule #1: human Nav1.7 in complex with hardwickii acid
| Supramolecule | Name: human Nav1.7 in complex with hardwickii acid / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 250 KDa |
-Macromolecule #1: Sodium channel protein type 9 subunit alpha
| Macromolecule | Name: Sodium channel protein type 9 subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 230.9225 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: WSHPQFEKGG GARGGSGGGS WSHPQFEKGF DYKDDDDKGT MAMLPPPGPQ SFVHFTKQSL ALIEQRIAER KSKEPKEEKK DDDEEAPKP SSDLEAGKQL PFIYGDIPPG MVSEPLEDLD PYYADKKTFI VLNKGKTIFR FNATPALYML SPFSPLRRIS I KILVHSLF ...String: WSHPQFEKGG GARGGSGGGS WSHPQFEKGF DYKDDDDKGT MAMLPPPGPQ SFVHFTKQSL ALIEQRIAER KSKEPKEEKK DDDEEAPKP SSDLEAGKQL PFIYGDIPPG MVSEPLEDLD PYYADKKTFI VLNKGKTIFR FNATPALYML SPFSPLRRIS I KILVHSLF SMLIMCTILT NCIFMTMNNP PDWTKNVEYT FTGIYTFESL VKILARGFCV GEFTFLRDPW NWLDFVVIVF AY LTEFVNL GNVSALRTFR VLRALKTISV IPGLKTIVGA LIQSVKKLSD VMILTVFCLS VFALIGLQLF MGNLKHKCFR NSL ENNETL ESIMNTLESE EDFRKYFYYL EGSKDALLCG FSTDSGQCPE GYTCVKIGRN PDYGYTSFDT FSWAFLALFR LMTQ DYWEN LYQQTLRAAG KTYMIFFVVV IFLGSFYLIN LILAVVAMAY EEQNQANIEE AKQKELEFQQ MLDRLKKEQE EAEAI AAAA AEYTSIRRSR IMGLSESSSE TSKLSSKSAK ERRNRRKKKN QKKLSSGEEK GDAEKLSKSE SEDSIRRKSF HLGVEG HRR AHEKRLSTPN QSPLSIRGSL FSARRSSRTS LFSFKGRGRD IGSETEFADD EHSIFGDNES RRGSLFVPHR PQERRSS NI SQASRSPPML PVNGKMHSAV DCNGVVSLVD GRSALMLPNG QLLPEVIIDK ATSDDSGTTN QIHKKRRCSS YLLSEDML N DPNLRQRAMS RASILTNTVE ELEESRQKCP PWWYRFAHKF LIWNCSPYWI KFKKCIYFIV MDPFVDLAIT ICIVLNTLF MAMEHHPMTE EFKNVLAIGN LVFTGIFAAE MVLKLIAMDP YEYFQVGWNI FDSLIVTLSL VELFLADVEG LSVLRSFRLL RVFKLAKSW PTLNMLIKII GNSVGALGNL TLVLAIIVFI FAVVGMQLFG KSYKECVCKI NDDCTLPRWH MNDFFHSFLI V FRVLCGEW IETMWDCMEV AGQAMCLIVY MMVMVIGNLV VLNLFLALLL SSFSSDNLTA IEEDPDANNL QIAVTRIKKG IN YVKQTLR EFILKAFSKK PKISREIRQA EDLNTKKENY ISNHTLAEMS KGHNFLKEKD KISGFGSSVD KHLMEDSDGQ SFI HNPSLT VTVPIAPGES DLENMNAEEL SSDSDSEYSK VRLNRSSSSE CSTVDNPLPG EGEEAEAEPM NSDEPEACFT DGCV WRFSC CQVNIESGKG KIWWNIRKTC YKIVEHSWFE SFIVLMILLS SGALAFEDIY IERKKTIKII LEYADKIFTY IFILE MLLK WIAYGYKTYF TNAWCWLDFL IVDVSLVTLV ANTLGYSDLG PIKSLRTLRA LRPLRALSRF EGMRVVVNAL IGAIPS IMN VLLVCLIFWL IFSIMGVNLF AGKFYECINT TDGSRFPASQ VPNRSECFAL MNVSQNVRWK NLKVNFDNVG LGYLSLL QV ATFKGWTIIM YAAVDSVNVD KQPKYEYSLY MYIYFVVFII FGSFFTLNLF IGVIIDNFNQ QKKKLGGQDI FMTEEQKK Y YNAMKKLGSK KPQKPIPRPG NKIQGCIFDL VTNQAFDISI MVLICLNMVT MMVEKEGQSQ HMTEVLYWIN VVFIILFTG ECVLKLISLR HYYFTVGWNI FDFVVVIISI VGMFLADLIE TYFVSPTLFR VIRLARIGRI LRLVKGAKGI RTLLFALMMS LPALFNIGL LLFLVMFIYA IFGMSNFAYV KKEDGINDMF NFETFGNSMI CLFQITTSAG WDGLLAPILN SKPPDCDPKK V HPGSSVEG DCGNPSVGIF YFVSYIIISF LVVVNMYIAV ILENFSVATE ESTEPLSEDD FEMFYEVWEK FDPDATQFIE FS KLSDFAA ALDPPLLIAK PNKVQLIAMD LPMVSGDRIH CLDILFAFTK RVLGESGEMD SLRSQMEERF MSANPSKVSY EPI TTTLKR KQEDVSATVI QRAYRRYRLR QNVKNISSIY IKDGDRDDDL LNKKDMAFDN VNENSSPEKT DATSSTTSPP SYDS VTKPD KEKYEQDRTE KEDKGKDSKE SKK UniProtKB: Sodium channel protein type 9 subunit alpha |
-Macromolecule #2: Sodium channel subunit beta-1
| Macromolecule | Name: Sodium channel subunit beta-1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.732115 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGRLLALVVG AALVSSACGG CVEVDSETEA VYGMTFKILC ISCKRRSETN AETFTEWTFR QKGTEEFVKI LRYENEVLQL EEDERFEGR VVWNGSRGTK DLQDLSIFIT NVTYNHSGDY ECHVYRLLFF ENYEHNTSVV KKIHIEVVDK ANRDMASIVS E IMMYVLIV ...String: MGRLLALVVG AALVSSACGG CVEVDSETEA VYGMTFKILC ISCKRRSETN AETFTEWTFR QKGTEEFVKI LRYENEVLQL EEDERFEGR VVWNGSRGTK DLQDLSIFIT NVTYNHSGDY ECHVYRLLFF ENYEHNTSVV KKIHIEVVDK ANRDMASIVS E IMMYVLIV VLTIWLVAEM IYCYKKIAAA TETAAQENAS EYLAITSESK ENCTGVQVAE UniProtKB: Sodium channel regulatory subunit beta-1 |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 5 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #5: (4~{a}~{R},5~{S},6~{R},8~{a}~{R})-5-[2-(furan-3-yl)ethyl]-5,6,8~{...
| Macromolecule | Name: (4~{a}~{R},5~{S},6~{R},8~{a}~{R})-5-[2-(furan-3-yl)ethyl]-5,6,8~{a}-trimethyl-3,4,4~{a},6,7,8-hexahydronaphthalene-1-carboxylic acid type: ligand / ID: 5 / Number of copies: 1 / Formula: UK0 |
|---|---|
| Molecular weight | Theoretical: 316.435 Da |
| Chemical component information |  ChemComp-UK0: |
-Macromolecule #6: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 6 / Number of copies: 3 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #7: (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]sp...
| Macromolecule | Name: (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en type: ligand / ID: 7 / Number of copies: 1 / Formula: 9Z9 |
|---|---|
| Molecular weight | Theoretical: 544.805 Da |
| Chemical component information |  ChemComp-9Z9: |
-Macromolecule #8: 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 8 / Number of copies: 10 / Formula: LPE |
|---|---|
| Molecular weight | Theoretical: 510.708 Da |
| Chemical component information |  ChemComp-LPE: |
-Macromolecule #9: 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 9 / Number of copies: 4 / Formula: PCW |
|---|---|
| Molecular weight | Theoretical: 787.121 Da |
| Chemical component information |  ChemComp-PCW: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 220477 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)