+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM consensus map of the dimeric human CAF1-H3-H4 complex | |||||||||||||||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||||||||
 Keywords Keywords | Histone chaperone / Chromatin assembly factor / REPLICATION | |||||||||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||||||||||||||
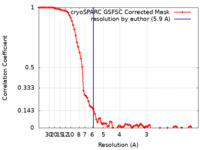
| Method | single particle reconstruction / cryo EM / Resolution: 5.9 Å | |||||||||||||||||||||||||||||||||
 Authors Authors | Liu CP / Yu ZY / Xu RM | |||||||||||||||||||||||||||||||||
| Funding support |  China, 10 items China, 10 items
| |||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Science / Year: 2023 Journal: Science / Year: 2023Title: Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1. Authors: Chao-Pei Liu / Zhenyu Yu / Jun Xiong / Jie Hu / Aoqun Song / Dongbo Ding / Cong Yu / Na Yang / Mingzhu Wang / Juan Yu / Peini Hou / Kangning Zeng / Zhenyu Li / Zhuqiang Zhang / Xinzheng ...Authors: Chao-Pei Liu / Zhenyu Yu / Jun Xiong / Jie Hu / Aoqun Song / Dongbo Ding / Cong Yu / Na Yang / Mingzhu Wang / Juan Yu / Peini Hou / Kangning Zeng / Zhenyu Li / Zhuqiang Zhang / Xinzheng Zhang / Wei Li / Zhiguo Zhang / Bing Zhu / Guohong Li / Rui-Ming Xu /   Abstract: Chromatin inheritance entails de novo nucleosome assembly after DNA replication by chromatin assembly factor-1 (CAF-1). Yet direct knowledge about CAF-1's histone binding mode and nucleosome assembly ...Chromatin inheritance entails de novo nucleosome assembly after DNA replication by chromatin assembly factor-1 (CAF-1). Yet direct knowledge about CAF-1's histone binding mode and nucleosome assembly process is lacking. In this work, we report the crystal structure of human CAF-1 in the absence of histones and the cryo-electron microscopy structure of CAF-1 in complex with histones H3 and H4. One histone H3-H4 heterodimer is bound by one CAF-1 complex mainly through the p60 subunit and the acidic domain of the p150 subunit. We also observed a dimeric CAF-1-H3-H4 supercomplex in which two H3-H4 heterodimers are poised for tetramer assembly and discovered that CAF-1 facilitates right-handed DNA wrapping of H3-H4 tetramers. These findings signify the involvement of DNA in H3-H4 tetramer formation and suggest a right-handed nucleosome precursor in chromatin replication. | |||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35708.map.gz emd_35708.map.gz | 30.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35708-v30.xml emd-35708-v30.xml emd-35708.xml emd-35708.xml | 17.4 KB 17.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35708_fsc.xml emd_35708_fsc.xml | 7.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_35708.png emd_35708.png | 57 KB | ||
| Others |  emd_35708_half_map_1.map.gz emd_35708_half_map_1.map.gz emd_35708_half_map_2.map.gz emd_35708_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35708 http://ftp.pdbj.org/pub/emdb/structures/EMD-35708 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35708 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35708 | HTTPS FTP |
-Validation report
| Summary document |  emd_35708_validation.pdf.gz emd_35708_validation.pdf.gz | 908.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35708_full_validation.pdf.gz emd_35708_full_validation.pdf.gz | 907.7 KB | Display | |
| Data in XML |  emd_35708_validation.xml.gz emd_35708_validation.xml.gz | 15.2 KB | Display | |
| Data in CIF |  emd_35708_validation.cif.gz emd_35708_validation.cif.gz | 20.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35708 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35708 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35708 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35708 | HTTPS FTP |
-Related structure data
| Related structure data |  7y5kC  7y5lC  7y5oC  7y5uC  7y5vC  7y5wC  7y60C  7y61C  8iqfC  8iqgC  8j6sC  8j6tC C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_35708.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35708.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||
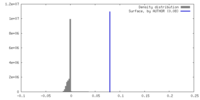
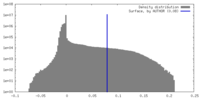
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_35708_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_35708_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Dimeric CAF1-H3-H4 complex
| Entire | Name: Dimeric CAF1-H3-H4 complex |
|---|---|
| Components |
|
-Supramolecule #1: Dimeric CAF1-H3-H4 complex
| Supramolecule | Name: Dimeric CAF1-H3-H4 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 523 KDa |
-Supramolecule #2: CAF-1
| Supramolecule | Name: CAF-1 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1, #4-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: H3-H4
| Supramolecule | Name: H3-H4 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 20 mM Hepes, pH 7.5, 50 mM NaCl and 1 mM DTT |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK I |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 321 / Target criteria: Correlation coefficient |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)