[English] 日本語
 Yorodumi
Yorodumi- EMDB-3568: 3D reconstruction for the thin CcdA-CcdB-DNA filaments based on t... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3568 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
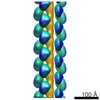
| Title | 3D reconstruction for the thin CcdA-CcdB-DNA filaments based on the negative stain electron microscopy dataset. | |||||||||
 Map data Map data | 3D reconstruction for the thin CcdA-CcdB-DNA filaments based on the negative stain electron microscopy dataset. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   | |||||||||
| Method | helical reconstruction / negative staining / Resolution: 23.0 Å | |||||||||
 Authors Authors | Vandervelde A / Efremov R / Loris R | |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2017 Journal: Nucleic Acids Res / Year: 2017Title: Molecular mechanism governing ratio-dependent transcription regulation in the ccdAB operon. Authors: Alexandra Vandervelde / Igor Drobnak / San Hadži / Yann G-J Sterckx / Thomas Welte / Henri De Greve / Daniel Charlier / Rouslan Efremov / Remy Loris / Jurij Lah /    Abstract: Bacteria can become transiently tolerant to several classes of antibiotics. This phenomenon known as persistence is regulated by small genetic elements called toxin-antitoxin modules with intricate ...Bacteria can become transiently tolerant to several classes of antibiotics. This phenomenon known as persistence is regulated by small genetic elements called toxin-antitoxin modules with intricate yet often poorly understood self-regulatory features. Here, we describe the structures of molecular complexes and interactions that drive the transcription regulation of the ccdAB toxin-antitoxin module. Low specificity and affinity of the antitoxin CcdA2 for individual binding sites on the operator are enhanced by the toxin CcdB2, which bridges the CcdA2 dimers. This results in a unique extended repressing complex that spirals around the operator and presents equally spaced DNA binding sites. The multivalency of binding sites induces a digital on-off switch for transcription, regulated by the toxin:antitoxin ratio. The ratio at which this switch occurs is modulated by non-specific interactions with the excess chromosomal DNA. Altogether, we present the molecular mechanisms underlying the ratio-dependent transcriptional regulation of the ccdAB operon. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3568.map.gz emd_3568.map.gz | 2.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3568-v30.xml emd-3568-v30.xml emd-3568.xml emd-3568.xml | 16.5 KB 16.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3568_fsc.xml emd_3568_fsc.xml | 2.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_3568.png emd_3568.png | 26.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3568 http://ftp.pdbj.org/pub/emdb/structures/EMD-3568 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3568 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3568 | HTTPS FTP |
-Validation report
| Summary document |  emd_3568_validation.pdf.gz emd_3568_validation.pdf.gz | 232.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3568_full_validation.pdf.gz emd_3568_full_validation.pdf.gz | 231.5 KB | Display | |
| Data in XML |  emd_3568_validation.xml.gz emd_3568_validation.xml.gz | 7.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3568 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3568 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3568 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3568 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3568.map.gz / Format: CCP4 / Size: 2.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3568.map.gz / Format: CCP4 / Size: 2.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D reconstruction for the thin CcdA-CcdB-DNA filaments based on the negative stain electron microscopy dataset. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.58 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Thin filaments formed as the complex between DNA, antitoxin CcdA ...
| Entire | Name: Thin filaments formed as the complex between DNA, antitoxin CcdA and toxin CcdB. |
|---|---|
| Components |
|
-Supramolecule #1: Thin filaments formed as the complex between DNA, antitoxin CcdA ...
| Supramolecule | Name: Thin filaments formed as the complex between DNA, antitoxin CcdA and toxin CcdB. type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: antitoxin CcdA and toxin CcdB
| Supramolecule | Name: antitoxin CcdA and toxin CcdB / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: DNA
| Supramolecule | Name: DNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Antitoxin CcdA
| Macromolecule | Name: Antitoxin CcdA / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MKQRITVTVD SDSYQLLKAY DVNISGLVST TMQNEARRLR AERWKAENQE GMAEVARFIE MNGSFADENR DW |
-Macromolecule #2: Toxin CcdB
| Macromolecule | Name: Toxin CcdB / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MQFKVYTYKR ESRYRLFVDV QSDIIDTPGR RMVIPLASAR LLSDKVSREL YPVVHIGDES WRMMTTDMAS VPVSVIGEEV ADLSHREND IKNAINLMFW GI |
-Macromolecule #3: DNA
| Macromolecule | Name: DNA / type: dna / ID: 3 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: AATTGT GAT GCTTCTAAAA TTACTAAAAT TGAGGATTTT AATGCTACAA CAATGCC TG CCTCTTCTTA TTTCTCCGGA GATCCGAAAA CCCCAAGTTA CGGATCTT C CTCTCCCTCC GCACAGCGTT ACATCCCGTC AGCACAGCAT GTAGTGCCT CATACAGTTG ...String: AATTGT GAT GCTTCTAAAA TTACTAAAAT TGAGGATTTT AATGCTACAA CAATGCC TG CCTCTTCTTA TTTCTCCGGA GATCCGAAAA CCCCAAGTTA CGGATCTT C CTCTCCCTCC GCACAGCGTT ACATCCCGTC AGCACAGCAT GTAGTGCCT CATACAGTTG CCCATGGCAC TATATGTTGT GTTGTATCTC TGGACTGTGA TGCGCCGCG CAGGGGCGGA AAACAGCGAT ATGATGATTT TCTCAGCGTT G TACACTTC CGGAAAGTCG TTTATTCAAA TAAAGTCGGA TCCATACGAA AC GGGAATG CGGTAATTAC GCTTTGTTTT TATAAGTCAG ATTTTAATTT TTA TTGGTT AACATAACGA AAGGTAAAAT ACATAAGGCT TACTAAAAGC CAGA TAACA GTATGCGTAT TTGCGCGCTG ATTTTTGCGG TATAAGAATA TATAC TGAT ATGTATACCC GAAGTATGTC AAAAAGAGGT GTGCTATGAA GCAGCG TAT TACAGTGACA GTTGACAGCG ACAGCTATCA GTTGCTCAAG GCATATG AT GTCAATATCT CCGGTCTGGT AAGCACAACC ATGCAGAATG AAGCCCGT C GTCTGCGTGC CGAACGCTGG AAAGCGGAAA ATCAGGAAGG GATGGCTGA GGTCGCCCGG TTTATTGAAA TGAACGGCTC TTTTGCTGAC GAGAACAGGG ACTGGTGAA ATGCAGTTTA AGGTTTACAC CTATAAAAGA GAGAGCCGTT A TCGTCTGT TTGTGGATGT ACAGAGTGAT ATTATTGACA CGCCCGGGCG AC GGATGGT GATCCCCCTG GCCAGTGCAC GTCTGCTGTC AGATAAAGTC TCC CGTGAA CTTTACCCGG TGGTGCATAT CGGGGATGAA AGCTGGCGCA TGAT GACCA CCGATATGGC CAGTGTGCCG GTCTCCGTTA TCGGGGAAGA AGTGG CTGA TCTCAGCCAC CGCGAAAATG ACATCAAAAA CGCCATTAAC C |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
Details: Just before staining, the sample was supplemented with 5 mM MgAc2 (final concentration). | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl formate Details: The sample was applied to a freshly glow-discharged grid for five minutes after which the grid was washed with 5 mM MgAc2 and adsorbed protein stained with 1% uranyl formate. | ||||||||
| Grid | Model: Electron Microscopy Sciences, continuous carbon/formvar Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.045 kPa / Details: ELMO glow discharger | ||||||||
| Details | The binding reactions occurred at 20degC. DNA: 15 nM CcdA: 2.5 uM (dimer concentration) CcdB: 2.5 uM (dimer concentration) DNA was first incubated with CcdA for 15 minutes. After adding CcdB, the resulting mixture was incubated for another 15 minutes before preparing the EM sample. |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 1400 |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Number grids imaged: 1 / Number real images: 358 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Calibrated defocus max: 3.0 µm / Calibrated defocus min: 1.2 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 3.4 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.8 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: JEOL |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















