[English] 日本語
 Yorodumi
Yorodumi- EMDB-35281: Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - State 5S RNP | |||||||||
 Map data Map data | local resolution filtered map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ribosome / Ribosome biogenesis / pre-60S ribosome | |||||||||
| Function / homology |  Function and homology information Function and homology informationdolichyl-diphosphooligosaccharide-protein glycotransferase / dolichyl-diphosphooligosaccharide-protein glycotransferase activity / rRNA (cytosine-C5-)-methyltransferase activity / intracellular mRNA localization / protein N-linked glycosylation via asparagine / PeBoW complex / rRNA primary transcript binding / rRNA base methylation / maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / preribosome, small subunit precursor ...dolichyl-diphosphooligosaccharide-protein glycotransferase / dolichyl-diphosphooligosaccharide-protein glycotransferase activity / rRNA (cytosine-C5-)-methyltransferase activity / intracellular mRNA localization / protein N-linked glycosylation via asparagine / PeBoW complex / rRNA primary transcript binding / rRNA base methylation / maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / preribosome, small subunit precursor / maturation of 5.8S rRNA / ribosomal large subunit binding / preribosome, large subunit precursor / mRNA transport / ribosomal subunit export from nucleus / ribonucleoprotein complex binding / maturation of LSU-rRNA / translation initiation factor activity / endomembrane system / post-translational protein modification / nuclear periphery / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / ribosome assembly / ribosomal large subunit biogenesis / cytosolic ribosome assembly / rRNA processing / ribosome biogenesis / large ribosomal subunit / ribosomal small subunit biogenesis / 5S rRNA binding / ribosomal large subunit assembly / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / cytoplasmic translation / RNA helicase activity / negative regulation of translation / rRNA binding / RNA helicase / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / mRNA binding / GTP binding / nucleolus / ATP hydrolysis activity / DNA binding / RNA binding / zinc ion binding / nucleoplasm / ATP binding / metal ion binding / nucleus / membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus) | |||||||||
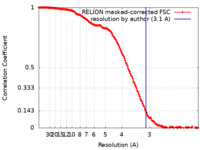
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Lau B / Huang Z / Beckmann R / Hurt E / Cheng J | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: EMBO Rep / Year: 2023 Journal: EMBO Rep / Year: 2023Title: Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis. Authors: Benjamin Lau / Zixuan Huang / Nikola Kellner / Shuangshuang Niu / Otto Berninghausen / Roland Beckmann / Ed Hurt / Jingdong Cheng /   Abstract: Ribosome biogenesis proceeds along a multifaceted pathway from the nucleolus to the cytoplasm that is extensively coupled to several quality control mechanisms. However, the mode by which 5S ...Ribosome biogenesis proceeds along a multifaceted pathway from the nucleolus to the cytoplasm that is extensively coupled to several quality control mechanisms. However, the mode by which 5S ribosomal RNA is incorporated into the developing pre-60S ribosome, which in humans links ribosome biogenesis to cell proliferation by surveillance by factors such as p53-MDM2, is poorly understood. Here, we report nine nucleolar pre-60S cryo-EM structures from Chaetomium thermophilum, one of which clarifies the mechanism of 5S RNP incorporation into the early pre-60S. Successive assembly states then represent how helicases Dbp10 and Spb4, and the Pumilio domain factor Puf6 act in series to surveil the gradual folding of the nearby 25S rRNA domain IV. Finally, the methyltransferase Spb1 methylates a universally conserved guanine nucleotide in the A-loop of the peptidyl transferase center, thereby licensing further maturation. Our findings provide insight into the hierarchical action of helicases in safeguarding rRNA tertiary structure folding and coupling to surveillance mechanisms that culminate in local RNA modification. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35281.map.gz emd_35281.map.gz | 163.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35281-v30.xml emd-35281-v30.xml emd-35281.xml emd-35281.xml | 85.4 KB 85.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35281_fsc.xml emd_35281_fsc.xml | 14.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_35281.png emd_35281.png | 117.6 KB | ||
| Filedesc metadata |  emd-35281.cif.gz emd-35281.cif.gz | 18.3 KB | ||
| Others |  emd_35281_additional_1.map.gz emd_35281_additional_1.map.gz emd_35281_additional_2.map.gz emd_35281_additional_2.map.gz emd_35281_half_map_1.map.gz emd_35281_half_map_1.map.gz emd_35281_half_map_2.map.gz emd_35281_half_map_2.map.gz | 246.9 MB 225.5 MB 225.7 MB 225.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35281 http://ftp.pdbj.org/pub/emdb/structures/EMD-35281 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35281 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35281 | HTTPS FTP |
-Validation report
| Summary document |  emd_35281_validation.pdf.gz emd_35281_validation.pdf.gz | 922.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35281_full_validation.pdf.gz emd_35281_full_validation.pdf.gz | 921.6 KB | Display | |
| Data in XML |  emd_35281_validation.xml.gz emd_35281_validation.xml.gz | 22.6 KB | Display | |
| Data in CIF |  emd_35281_validation.cif.gz emd_35281_validation.cif.gz | 30.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35281 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35281 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35281 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35281 | HTTPS FTP |
-Related structure data
| Related structure data |  8i9rMC  8i9pC  8i9tC  8i9vC  8i9wC  8i9xC  8i9yC  8i9zC  8ia0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35281.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35281.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | local resolution filtered map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: DeepEMhancer filtered map
| File | emd_35281_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer filtered map | ||||||||||||
| Projections & Slices |
| ||||||||||||
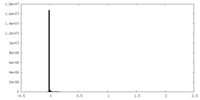
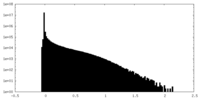
| Density Histograms |
-Additional map: without post processing
| File | emd_35281_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | without post processing | ||||||||||||
| Projections & Slices |
| ||||||||||||
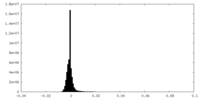
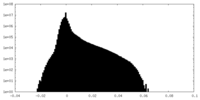
| Density Histograms |
-Half map: #2
| File | emd_35281_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_35281_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : pre-60S ribosome
+Supramolecule #1: pre-60S ribosome
+Supramolecule #2: ribosome
+Macromolecule #1: RNA (3341-MER)
+Macromolecule #2: RNA (256-MER)
+Macromolecule #47: RNA (119-MER)
+Macromolecule #3: Brix domain-containing protein
+Macromolecule #4: Ribosome biogenesis protein C8F11.04
+Macromolecule #5: Ribosome biogenesis protein ERB1
+Macromolecule #6: RNA helicase
+Macromolecule #7: Nucleolar GTP-binding protein 1
+Macromolecule #8: Putative RNA-binding protein
+Macromolecule #9: Pescadillo homolog
+Macromolecule #10: 60S ribosomal protein l7-like protein
+Macromolecule #11: Eukaryotic translation initiation factor 6
+Macromolecule #12: Ribosome biogenesis protein RLP24
+Macromolecule #13: Nucleolar protein 16
+Macromolecule #14: rRNA-processing protein EBP2
+Macromolecule #15: Ribosomal RNA-processing protein 15
+Macromolecule #16: 60S ribosomal protein L3-like protein
+Macromolecule #17: 60S ribosomal protein L4-like protein
+Macromolecule #18: 60S ribosomal protein L6
+Macromolecule #19: 60S ribosomal protein L8
+Macromolecule #20: 60S ribosomal protein L13
+Macromolecule #21: 60S ribosomal protein L14-like protein
+Macromolecule #22: Ribosomal protein L15
+Macromolecule #23: 60S ribosomal protein L16-like protein
+Macromolecule #24: 60S ribosomal protein l17-like protein
+Macromolecule #25: Ribosomal protein L18-like protein
+Macromolecule #26: 60S ribosomal protein L20
+Macromolecule #27: 60S ribosomal protein l21-like protein
+Macromolecule #28: 60S ribosomal protein l23-like protein
+Macromolecule #29: 60S ribosomal protein L26-like protein
+Macromolecule #30: 60S ribosomal protein L32-like protein
+Macromolecule #31: 60S ribosomal protein l33-like protein
+Macromolecule #32: dolichyl-diphosphooligosaccharide--protein glycotransferase
+Macromolecule #33: 60S ribosomal protein L36
+Macromolecule #34: Ribosomal protein L37
+Macromolecule #35: Ribosomal RNA-processing protein 1
+Macromolecule #36: Brix domain-containing protein
+Macromolecule #37: Protein MAK16
+Macromolecule #38: 60S ribosome biogenesis protein Rrp14
+Macromolecule #39: Ribosome production factor 2 homolog
+Macromolecule #40: Brix domain-containing protein
+Macromolecule #41: RNA methyltransferase nop2-like protein
+Macromolecule #42: 60S ribosome subunit biogenesis protein NIP7
+Macromolecule #43: Ribosomal protein
+Macromolecule #44: Ribosome biogenesis regulatory protein
+Macromolecule #45: Putative ribosomal protein
+Macromolecule #46: 60S ribosomal protein l5-like protein
+Macromolecule #48: 60S ribosomal subunit-like protein
+Macromolecule #49: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 44.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)