[English] 日本語
 Yorodumi
Yorodumi- EMDB-35201: Cryo-EM structure of Pseudomonas aeruginosa FtsE(WT)X/EnvC comple... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Pseudomonas aeruginosa FtsE(WT)X/EnvC complex in peptidisc | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Type VII ABC transporter / divisome / PG hydrolysis / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationseptum digestion after cytokinesis / cell division site / transmembrane transporter activity / metalloendopeptidase activity / cell division / ATP hydrolysis activity / ATP binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
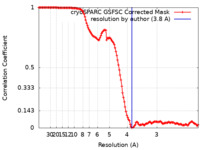
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Xu X / Li J / Luo M | |||||||||
| Funding support |  Singapore, 1 items Singapore, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Mechanistic insights into the regulation of cell wall hydrolysis by FtsEX and EnvC at the bacterial division site. Authors: Xin Xu / Jianwei Li / Wan-Zhen Chua / Martin A Pages / Jian Shi / Juan A Hermoso / Thomas Bernhardt / Lok-To Sham / Min Luo /    Abstract: The peptidoglycan (PG) cell wall produced by the bacterial division machinery is initially shared between the daughters and must be split to promote cell separation and complete division. In gram- ...The peptidoglycan (PG) cell wall produced by the bacterial division machinery is initially shared between the daughters and must be split to promote cell separation and complete division. In gram-negative bacteria, enzymes that cleave PG called amidases play major roles in the separation process. To prevent spurious cell wall cleavage that can lead to cell lysis, amidases like AmiB are autoinhibited by a regulatory helix. Autoinhibition is relieved at the division site by the activator EnvC, which is in turn regulated by the ATP-binding cassette (ABC) transporter-like complex called FtsEX. EnvC is also known to be autoinhibited by a regulatory helix (RH), but how its activity is modulated by FtsEX and the mechanism by which it activates the amidases have remained unclear. Here, we investigated this regulation by determining the structure of FtsEX alone with or without bound ATP, in complex with EnvC, and in a FtsEX-EnvC-AmiB supercomplex. In combination with biochemical studies, the structures reveal that ATP binding is likely to activate FtsEX-EnvC and promote its association with AmiB. Furthermore, the AmiB activation mechanism is shown to involve a RH rearrangement. In the activated state of the complex, the inhibitory helix of EnvC is released, freeing it to associate with the RH of AmiB, which liberates its active site for PG cleavage. These regulatory helices are found in many EnvC proteins and amidases throughout gram-negative bacteria, suggesting that the activation mechanism is broadly conserved and a potential target for lysis-inducing antibiotics that misregulate the complex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35201.map.gz emd_35201.map.gz | 204 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35201-v30.xml emd-35201-v30.xml emd-35201.xml emd-35201.xml | 16.7 KB 16.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35201_fsc.xml emd_35201_fsc.xml | 14.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_35201.png emd_35201.png | 25.7 KB | ||
| Filedesc metadata |  emd-35201.cif.gz emd-35201.cif.gz | 6 KB | ||
| Others |  emd_35201_half_map_1.map.gz emd_35201_half_map_1.map.gz emd_35201_half_map_2.map.gz emd_35201_half_map_2.map.gz | 200.7 MB 200.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35201 http://ftp.pdbj.org/pub/emdb/structures/EMD-35201 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35201 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35201 | HTTPS FTP |
-Related structure data
| Related structure data |  8i6oMC  8i6qC  8i6rC  8i6sC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35201.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35201.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.105 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35201_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
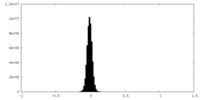
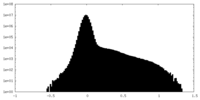
| Density Histograms |
-Half map: #1
| File | emd_35201_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
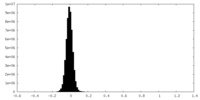
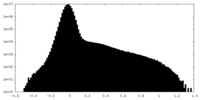
| Density Histograms |
- Sample components
Sample components
-Entire : FtsEX-EnvC
| Entire | Name: FtsEX-EnvC |
|---|---|
| Components |
|
-Supramolecule #1: FtsEX-EnvC
| Supramolecule | Name: FtsEX-EnvC / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #2, #1, #3 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 169 KDa |
-Macromolecule #1: Cell division protein FtsX
| Macromolecule | Name: Cell division protein FtsX / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 36.964359 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSANDLPRGP EEGAPERKTR EKPSQEQTDW SGSFSAYLES HRASLVDSLR RLFGHPFGSF FTCLVMGITL SLPMGLSLLL NNVERLGGS WQRAAQISLF LDLKTSENQG QDLREQIERL PDVIEAQLIS REQALSELQE QSGLGEALKE LPENPLPPVI S VTPKQIDR ...String: MSANDLPRGP EEGAPERKTR EKPSQEQTDW SGSFSAYLES HRASLVDSLR RLFGHPFGSF FTCLVMGITL SLPMGLSLLL NNVERLGGS WQRAAQISLF LDLKTSENQG QDLREQIERL PDVIEAQLIS REQALSELQE QSGLGEALKE LPENPLPPVI S VTPKQIDR AGLEALRQQL AELPHVQQAQ LDLVWVERLS AILKLGERFV FGLTILLVLT LLLVVGNTIR LHIENRRNEI EV IKLVGGT DGYVRRPFLY MGALYGLGAG ILSWALLAYS LNWLNGSVVN LSGLYGSDFG LQGVPLDDGL SLTVGAVLLG WVG AWLAVA RHLRELAPR UniProtKB: Cell division protein FtsX |
-Macromolecule #2: Cell division ATP-binding protein FtsE
| Macromolecule | Name: Cell division ATP-binding protein FtsE / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.649666 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIRFEQVGKR YPNGHVGLHE VSFRVHRGEI LFVTGHSGAG KSTLLRLILA MERPTSGKLL LGGQDLGRIT TAQIPFLRRQ IGVVFQNHQ LLTDRTVADN IALPLQILGM PKPEIAKRVA SALERVNLKE KGEALPSDLS TGQQQRVGIA RAIVHQPALL L ADEPTGNL ...String: MIRFEQVGKR YPNGHVGLHE VSFRVHRGEI LFVTGHSGAG KSTLLRLILA MERPTSGKLL LGGQDLGRIT TAQIPFLRRQ IGVVFQNHQ LLTDRTVADN IALPLQILGM PKPEIAKRVA SALERVNLKE KGEALPSDLS TGQQQRVGIA RAIVHQPALL L ADEPTGNL DPRLASEIMG VFEDINRLGT TVLIASHDLA LIARMRHRML TLQRGRIIAD REDEA UniProtKB: Cell division ATP-binding protein FtsE |
-Macromolecule #3: Membrane-bound metallopeptidase
| Macromolecule | Name: Membrane-bound metallopeptidase / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.353684 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DERADTQRQL EQTQKDIGEL KKLLDGIQQE KSGVQKQLKS TETEMGDLEK QIKALQDELD KSEAELKRLD GEKKKLQDAR IEQQRLLAI QARAAYQSGR EEYLKLLLNQ EHPEKFSRTL TYYDYINKAR LEQLASFNET LRQLANVEQD ISAQKAEQLS K QGELDSRR ...String: DERADTQRQL EQTQKDIGEL KKLLDGIQQE KSGVQKQLKS TETEMGDLEK QIKALQDELD KSEAELKRLD GEKKKLQDAR IEQQRLLAI QARAAYQSGR EEYLKLLLNQ EHPEKFSRTL TYYDYINKAR LEQLASFNET LRQLANVEQD ISAQKAEQLS K QGELDSRR EALAATRKER QQALAKLNSD YRERDQKLKS RQQDQAELAK VLRTIEETLA RQAREAAAAA ERERQRALAA ER ERARQQQ AAPGRVTSPP REPAPGPLVS STGAVYGGAF GSARGKLPWP VNGRVVARFG SQRGDDPRAK WDGVLISASA GST VRAVHG GRVVFADWLR GAGLLVILDH GGGYLSLYGH NQSLLKDAGD TVKAGDPIAT VGTSGGQSSP AVYFAIRHQG RPAD PTTWC RAQG UniProtKB: Membrane-bound metallopeptidase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 25 mM Tris, pH 7.5, 150 mM NaCl |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)