+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Acyl-ACP synthetase structure bound to AMP-MC7 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Acyl-ACP synthetase / Tool enzyme / CYTOSOLIC PROTEIN | |||||||||
| Function / homology | ligase activity, forming carbon-sulfur bonds / : / ANL, N-terminal domain / AMP-binding enzyme C-terminal domain / AMP-binding enzyme, C-terminal domain / AMP-dependent synthetase/ligase / AMP-binding enzyme / AMP-binding enzyme, C-terminal domain superfamily / Acyl-acyl carrier protein synthetase Function and homology information Function and homology information | |||||||||
| Biological species |  Vibrio harveyi (bacteria) Vibrio harveyi (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.76 Å | |||||||||
 Authors Authors | Huang H / Wang C / Chang S / Cui T / Xu Y / Zhang H / Zhou C / Zhang X / Feng Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2025 Journal: Nat Struct Mol Biol / Year: 2025Title: Structure and catalytic mechanism of exogenous fatty acid recycling by AasS, a versatile acyl-ACP synthetase. Authors: Haomin Huang / Chen Wang / Shenghai Chang / Tao Cui / Yongchang Xu / Man Huang / Huimin Zhang / Chun Zhou / Xing Zhang / Youjun Feng /   Abstract: Fatty acids (FAs) are essential building blocks for all the domains of life, of which bacterial de novo synthesis, called type II FA synthesis (FAS II), is energetically expensive. The recycling of ...Fatty acids (FAs) are essential building blocks for all the domains of life, of which bacterial de novo synthesis, called type II FA synthesis (FAS II), is energetically expensive. The recycling of exogenous FAs (eFAs) partially relieves the FAS II demand and, therefore, compromises the efficacy of FAS II-directed antimicrobials. The versatile acyl-acyl carrier protein (ACP) synthetase, AasS, enables bacterial channeling of diverse eFA nutrients through holo-ACP, an activated form of ACP. However, the molecular mechanism for AasS catalysis is not fully understood. Here we report a series of cryo-electron microscopy structures of AasS from the bioluminescent bacterium Vibrio harveyi to provide insights into the catalytic cycle. AasS forms a ring-shaped hexamer, with each protomer folding into two distinct domains. Biochemical and structural analysis suggests that AasS accommodates distinct eFA substrates and the conserved W230 residue has a gating role. Adenosine triphosphate and Mg binding converts the AasS hexamer to a tetramer, which is likely needed for the acyl adenylate intermediate formation. Afterward, AasS reverts to the hexamer conformation in adaption to acyl-ACP production. The complete landscape for eFA scavenging lays a foundation for exploiting the versatility of AasS in biopharmaceuticals. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35190.map.gz emd_35190.map.gz | 118 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35190-v30.xml emd-35190-v30.xml emd-35190.xml emd-35190.xml | 14.6 KB 14.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35190.png emd_35190.png | 153.2 KB | ||
| Filedesc metadata |  emd-35190.cif.gz emd-35190.cif.gz | 5.7 KB | ||
| Others |  emd_35190_half_map_1.map.gz emd_35190_half_map_1.map.gz emd_35190_half_map_2.map.gz emd_35190_half_map_2.map.gz | 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35190 http://ftp.pdbj.org/pub/emdb/structures/EMD-35190 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35190 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35190 | HTTPS FTP |
-Validation report
| Summary document |  emd_35190_validation.pdf.gz emd_35190_validation.pdf.gz | 876.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35190_full_validation.pdf.gz emd_35190_full_validation.pdf.gz | 875.8 KB | Display | |
| Data in XML |  emd_35190_validation.xml.gz emd_35190_validation.xml.gz | 14.2 KB | Display | |
| Data in CIF |  emd_35190_validation.cif.gz emd_35190_validation.cif.gz | 16.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35190 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35190 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35190 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35190 | HTTPS FTP |
-Related structure data
| Related structure data |  8i51MC  8i35C  8i3iC  8i49C  8i6mC  8i8dC  8i8eC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35190.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35190.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.014 Å | ||||||||||||||||||||||||||||||||||||
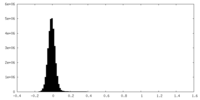
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35190_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
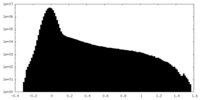
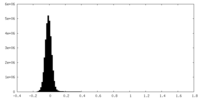
| Density Histograms |
-Half map: #1
| File | emd_35190_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
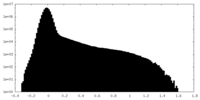
| Density Histograms |
- Sample components
Sample components
-Entire : Acyl-ACP Synthetase bound to AMP-MC7
| Entire | Name: Acyl-ACP Synthetase bound to AMP-MC7 |
|---|---|
| Components |
|
-Supramolecule #1: Acyl-ACP Synthetase bound to AMP-MC7
| Supramolecule | Name: Acyl-ACP Synthetase bound to AMP-MC7 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Vibrio harveyi (bacteria) Vibrio harveyi (bacteria) |
-Macromolecule #1: Acyl-acyl carrier protein synthetase
| Macromolecule | Name: Acyl-acyl carrier protein synthetase / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio harveyi (bacteria) Vibrio harveyi (bacteria) |
| Molecular weight | Theoretical: 60.496258 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNQYVNDPSN YQLLIKNLLF SPVAFNPEQE IVYANHRRHS YKTFHDRVRQ FANALTKMGV KKGDTVAVMD YDSHRYLECY FAIPMIGAK LHMINVRLSP EQILYTIDHA EDDIILIHEE FLPILDQIKG RIDTVTRYVV LRDDEECEYE RLLEQESTEY N FPDFDENT ...String: MNQYVNDPSN YQLLIKNLLF SPVAFNPEQE IVYANHRRHS YKTFHDRVRQ FANALTKMGV KKGDTVAVMD YDSHRYLECY FAIPMIGAK LHMINVRLSP EQILYTIDHA EDDIILIHEE FLPILDQIKG RIDTVTRYVV LRDDEECEYE RLLEQESTEY N FPDFDENT VATTFYTTGT TGFPKGVFFT HRQLVLHTMG ILSTIGTNAS QGRLHQGDIY MPITPMFHVH AWGLPYMATM LG VKQVYPG KYVPDVLLNL IEQEKVTFSH CVPTILHLLL SSPKSKAMDF SGWKVVIGGA ALPKALCKSA LERDIDVFAG YGM SETGPI LSIVQLTPEQ LELDVDQQAE YRSKTGKKVA LVEAYIVDED MNKLPHDGET AGEIVVRAPW LTPNYYKDNK NSKA LWRGG YLHTGDVAHI DDEGFIKITD RVKDMIKISG EWVSSLELED ILHQHQSVSE VAVIGMPHNK WGEVPLALVT LKEDA QVTE KELLGFAKDF INKGILAREA LLLKVKIVDE IAKTSVGKVD KKELRKLHL UniProtKB: Acyl-acyl carrier protein synthetase |
-Macromolecule #2: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 2 / Number of copies: 6 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #3: ADENOSINE MONOPHOSPHATE
| Macromolecule | Name: ADENOSINE MONOPHOSPHATE / type: ligand / ID: 3 / Number of copies: 6 / Formula: AMP |
|---|---|
| Molecular weight | Theoretical: 347.221 Da |
| Chemical component information |  ChemComp-AMP: |
-Macromolecule #4: 7-methoxy-7-oxidanylidene-heptanoic acid
| Macromolecule | Name: 7-methoxy-7-oxidanylidene-heptanoic acid / type: ligand / ID: 4 / Number of copies: 6 / Formula: OP3 |
|---|---|
| Molecular weight | Theoretical: 174.194 Da |
| Chemical component information |  ChemComp-OP3: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.76 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 199919 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)