[English] 日本語
 Yorodumi
Yorodumi- EMDB-34840: Bi-functional malonyl-CoA reductuase from Chloroflexus aurantiacus -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Bi-functional malonyl-CoA reductuase from Chloroflexus aurantiacus | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | malonyl-CoA reductase (MCR) / bi-functional enzyme / NADPH-dependent reduction / OXIDOREDUCTASE | |||||||||
| Function / homology | fatty acid elongation / oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor / short chain dehydrogenase / Enoyl-(Acyl carrier protein) reductase / Short-chain dehydrogenase/reductase SDR / NAD(P)-binding domain superfamily / nucleotide binding / metal ion binding / Short-chain dehydrogenase/reductase SDR Function and homology information Function and homology information | |||||||||
| Biological species |   Chloroflexus aurantiacus (bacteria) Chloroflexus aurantiacus (bacteria) | |||||||||
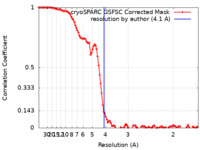
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | Ahn JW / Kim S | |||||||||
| Funding support |  Korea, Republic Of, 2 items Korea, Republic Of, 2 items
| |||||||||
 Citation Citation |  Journal: Int J Biol Macromol / Year: 2023 Journal: Int J Biol Macromol / Year: 2023Title: Cryo-EM structure of bifunctional malonyl-CoA reductase from Chloroflexus aurantiacus reveals a dynamic domain movement for high enzymatic activity. Authors: Jae-Woo Ahn / Sangwoo Kim / Jiyeon Hong / Kyung-Jin Kim /  Abstract: The platform chemical 3-hydroxypropionic acid is used to synthesize various valuable materials, including bioplastics. Bifunctional malonyl-CoA reductase is a key enzyme in 3-hydroxypropionic acid ...The platform chemical 3-hydroxypropionic acid is used to synthesize various valuable materials, including bioplastics. Bifunctional malonyl-CoA reductase is a key enzyme in 3-hydroxypropionic acid biosynthesis as it catalyzes the two-step reduction of malonyl-CoA to malonate semialdehyde to 3-hydroxypropionic acid. Here, we report the cryo-EM structure of a full-length malonyl-CoA reductase protein from Chloroflexus aurantiacus (CaMCR). The EM model of CaMCR reveals a tandem helix architecture comprising an N-terminal (CaMCR) and a C-terminal (CaMCR) domain. The CaMCR model also revealed that the enzyme undergoes a dynamic domain movement between CaMCR and CaMCR due to the presence of a flexible linker between these two domains. Increasing the flexibility and extension of the linker resulted in a twofold increase in enzyme activity, indicating that for CaMCR, domain movement is crucial for high enzyme activity. We also describe the structural features of CaMCR and CaMCR. This study reveals the protein structures underlying the molecular mechanism of CaMCR and thereby provides valuable information for future enzyme engineering to improve the productivity of 3-hydroxypropionic acid. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34840.map.gz emd_34840.map.gz | 113 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34840-v30.xml emd-34840-v30.xml emd-34840.xml emd-34840.xml | 19.2 KB 19.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34840_fsc.xml emd_34840_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_34840.png emd_34840.png | 142.8 KB | ||
| Masks |  emd_34840_msk_1.map emd_34840_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-34840.cif.gz emd-34840.cif.gz | 6.6 KB | ||
| Others |  emd_34840_half_map_1.map.gz emd_34840_half_map_1.map.gz emd_34840_half_map_2.map.gz emd_34840_half_map_2.map.gz | 115.7 MB 115.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34840 http://ftp.pdbj.org/pub/emdb/structures/EMD-34840 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34840 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34840 | HTTPS FTP |
-Related structure data
| Related structure data |  8hjwMC  8i6zC  8i70C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34840.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34840.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_34840_msk_1.map emd_34840_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_34840_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_34840_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Dimeric state of malonyl-CoA reductase (MCR) from Chloroflexus au...
| Entire | Name: Dimeric state of malonyl-CoA reductase (MCR) from Chloroflexus aurantiacus |
|---|---|
| Components |
|
-Supramolecule #1: Dimeric state of malonyl-CoA reductase (MCR) from Chloroflexus au...
| Supramolecule | Name: Dimeric state of malonyl-CoA reductase (MCR) from Chloroflexus aurantiacus type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Chloroflexus aurantiacus (bacteria) Chloroflexus aurantiacus (bacteria) |
| Molecular weight | Theoretical: 264 KDa |
-Macromolecule #1: Short-chain dehydrogenase/reductase SDR
| Macromolecule | Name: Short-chain dehydrogenase/reductase SDR / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Chloroflexus aurantiacus (bacteria) Chloroflexus aurantiacus (bacteria) |
| Molecular weight | Theoretical: 132.134219 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGTGRLAGK IALITGGAGN IGSELTRRFL AEGATVIISG RNRAKLTALA ERMQAEAGVP AKRIDLEVMD GSDPVAVRAG IEAIVARHG QIDILVNNAG SAGAQRRLAE IPLTEAELGP GAEETLHASI ANLLGMGWHL MRIAAPHMPV GSAVINVSTI F SRAEYYGR ...String: MSGTGRLAGK IALITGGAGN IGSELTRRFL AEGATVIISG RNRAKLTALA ERMQAEAGVP AKRIDLEVMD GSDPVAVRAG IEAIVARHG QIDILVNNAG SAGAQRRLAE IPLTEAELGP GAEETLHASI ANLLGMGWHL MRIAAPHMPV GSAVINVSTI F SRAEYYGR IPYVTPKAAL NALSQLAARE LGARGIRVNT IFPGPIESDR IRTVFQRMDQ LKGRPEGDTA HHFLNTMRLC RA NDQGALE RRFPSVGDVA DAAVFLASAE SAALSGETIE VTHGMELPAC SETSLLARTD LRTIDASGRT TLICAGDQIE EVM ALTGML RTCGSEVIIG FRSAAALAQF EQAVNESRRL AGADFTPPIA LPLDPRDPAT IDAVFDWAGE NTGGIHAAVI LPAT SHEPA PCVIEVDDER VLNFLADEIT GTIVIASRLA RYWQSQRLTP GARARGPRVI FLSNGADQNG NVYGRIQSAA IGQLI RVWR HEAELDYQRA SAAGDHVLPP VWANQIVRFA NRSLEGLEFA CAWTAQLLHS QRHINEITLN IPANISATTG ARSASV GWA ESLIGLHLGK VALITGGSAG IGGQIGRLLA LSGARVMLAA RDRHKLEQMQ AMIQSELAEV GYTDVEDRVH IAPGCDV SS EAQLADLVER TLSAFGTVDY LINNAGIAGV EEMVIDMPVE GWRHTLFANL ISNYSLMRKL APLMKKQGSG YILNVSSY F GGEKDAAIPY PNRADYAVSK AGQRAMAEVF ARFLGPEIQI NAIAPGPVEG DRLRGTGERP GLFARRARLI LENKRLNEL HAALIAAART DERSMHELVE LLLPNDVAAL EQNPAAPTAL RELARRFRSE GDPAASSSSA LLNRSIAAKL LARLHNGGYV LPADIFANL PNPPDPFFTR AQIDREARKV RDGIMGMLYL QRMPTEFDVA MATVYYLADR NVSGETFHPS GGLRYERTPT G GELFGLPS PERLAELVGS TVYLIGEHLT EHLNLLARAY LERYGARQVV MIVETETGAE TMRRLLHDHV EAGRLMTIVA GD QIEAAID QAITRYGRPG PVVCTPFRPL PTVPLVGRKD SDWSTVLSEA EFAELCEHQL THHFRVARKI ALSDGASLAL VTP ETTATS TTEQFALANF IKTTLHAFTA TIGVESERTA QRILINQVDL TRRARAEEPR DPHERQQELE RFIEAVLLVT APLP PEADT RYAGRIHRGR AITV UniProtKB: Short-chain dehydrogenase/reductase SDR |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 20mM Tris-HCl, 150mM NaCl |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.1 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)