+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 Omicron BA.1 Spike in complex with IY-2A | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Spike / Omicron variant / BA.1 / RBD / monoclonal antibody / Fab / IY-2A / class 4 / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationMaturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell ...Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell / membrane fusion / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / receptor ligand activity / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.28 Å | |||||||||
 Authors Authors | Chen X / Mohapatra A / Wu Y-M | |||||||||
| Funding support |  Taiwan, 1 items Taiwan, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural basis for a conserved neutralization epitope on the receptor-binding domain of SARS-CoV-2. Authors: Kuan-Ying A Huang / Xiaorui Chen / Arpita Mohapatra / Hong Thuy Vy Nguyen / Lisa Schimanski / Tiong Kit Tan / Pramila Rijal / Susan K Vester / Rory A Hills / Mark Howarth / Jennifer R Keeffe ...Authors: Kuan-Ying A Huang / Xiaorui Chen / Arpita Mohapatra / Hong Thuy Vy Nguyen / Lisa Schimanski / Tiong Kit Tan / Pramila Rijal / Susan K Vester / Rory A Hills / Mark Howarth / Jennifer R Keeffe / Alexander A Cohen / Leesa M Kakutani / Yi-Min Wu / Md Shahed-Al-Mahmud / Yu-Chi Chou / Pamela J Bjorkman / Alain R Townsend / Che Ma /    Abstract: Antibody-mediated immunity plays a crucial role in protection against SARS-CoV-2 infection. We isolated a panel of neutralizing anti-receptor-binding domain (RBD) antibodies elicited upon natural ...Antibody-mediated immunity plays a crucial role in protection against SARS-CoV-2 infection. We isolated a panel of neutralizing anti-receptor-binding domain (RBD) antibodies elicited upon natural infection and vaccination and showed that they recognize an immunogenic patch on the internal surface of the core RBD, which faces inwards and is hidden in the "down" state. These antibodies broadly neutralize wild type (Wuhan-Hu-1) SARS-CoV-2, Beta and Delta variants and some are effective against other sarbecoviruses. We observed a continuum of partially overlapping antibody epitopes from lower to upper part of the inner face of the RBD and some antibodies extend towards the receptor-binding motif. The majority of antibodies are substantially compromised by three mutational hotspots (S371L/F, S373P and S375F) in the lower part of the Omicron BA.1, BA.2 and BA.4/5 RBD. By contrast, antibody IY-2A induces a partial unfolding of this variable region and interacts with a conserved conformational epitope to tolerate all antigenic variations and neutralize diverse sarbecoviruses as well. This finding establishes that antibody recognition is not limited to the normal surface structures on the RBD. In conclusion, the delineation of functionally and structurally conserved RBD epitopes highlights potential vaccine and therapeutic candidates for COVID-19. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34808.map.gz emd_34808.map.gz | 121.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34808-v30.xml emd-34808-v30.xml emd-34808.xml emd-34808.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34808.png emd_34808.png | 91 KB | ||
| Filedesc metadata |  emd-34808.cif.gz emd-34808.cif.gz | 7.1 KB | ||
| Others |  emd_34808_half_map_1.map.gz emd_34808_half_map_1.map.gz emd_34808_half_map_2.map.gz emd_34808_half_map_2.map.gz | 102.2 MB 102.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34808 http://ftp.pdbj.org/pub/emdb/structures/EMD-34808 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34808 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34808 | HTTPS FTP |
-Related structure data
| Related structure data |  8hhzMC  7yckC  7yclC  7ycnC  8hhxC  8hhyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34808.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34808.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
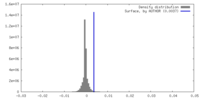
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34808_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
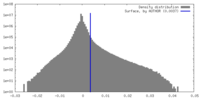
| Density Histograms |
-Half map: #1
| File | emd_34808_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of Delta Spike and IY-2A with two Fabs bound to one Spike...
| Entire | Name: Complex of Delta Spike and IY-2A with two Fabs bound to one Spike in 3-RBD-up conformation |
|---|---|
| Components |
|
-Supramolecule #1: Complex of Delta Spike and IY-2A with two Fabs bound to one Spike...
| Supramolecule | Name: Complex of Delta Spike and IY-2A with two Fabs bound to one Spike in 3-RBD-up conformation type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 550 KDa |
-Supramolecule #2: SARS-CoV-2 Delta Spike protein
| Supramolecule | Name: SARS-CoV-2 Delta Spike protein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: IY-2A
| Supramolecule | Name: IY-2A / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 140.204906 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKVKLLVLLC TFTATYAGTQ CVNLTTRTQL PPAYTNSFTR GVYYPDKVFR SSVLHSTQDL FLPFFSNVTW FHVISGTNGT KRFDNPVLP FNDGVYFASI EKSNIIRGWI FGTTLDSKTQ SLLIVNNATN VVIKVCEFQF CNDPFLDHKN NKSWMESEFR V YSSANNCT ...String: MKVKLLVLLC TFTATYAGTQ CVNLTTRTQL PPAYTNSFTR GVYYPDKVFR SSVLHSTQDL FLPFFSNVTW FHVISGTNGT KRFDNPVLP FNDGVYFASI EKSNIIRGWI FGTTLDSKTQ SLLIVNNATN VVIKVCEFQF CNDPFLDHKN NKSWMESEFR V YSSANNCT FEYVSQPFLM DLEGKQGNFK NLREFVFKNI DGYFKIYSKH TPIIVREPED LPQGFSALEP LVDLPIGINI TR FQTLLAL HRSYLTPGDS SSGWTAGAAA YYVGYLQPRT FLLKYNENGT ITDAVDCALD PLSETKCTLK SFTVEKGIYQ TSN FRVQPT ESIVRFPNIT NLCPFDEVFN ATRFASVYAW NRKRISNCVA DYSVLYNLAP FFTFKCYGVS PTKLNDLCFT NVYA DSFVI RGDEVRQIAP GQTGNIADYN YKLPDDFTGC VIAWNSNKLD SKVSGNYNYL YRLFRKSNLK PFERDISTEI YQAGN KPCN GVAGFNCYFP LRSYSFRPTY GVGHQPYRVV VLSFELLHAP ATVCGPKKST NLVKNKCVNF NFNGLKGTGV LTESNK KFL PFQQFGRDIA DTTDAVRDPQ TLEILDITPC SFGGVSVITP GTNTSNQVAV LYQGVNCTEV PVAIHADQLT PTWRVYS TG SNVFQTRAGC LIGAEYVNNS YECDIPIGAG ICASYQTQTK SHGSASSVAS QSIIAYTMSL GAENSVAYSN NSIAIPTN F TISVTTEILP VSMTKTSVDC TMYICGDSTE CSNLLLQYGS FCTQLKRALT GIAVEQDKNT QEVFAQVKQI YKTPPIKYF GGFNFSQILP DPSKPSKRSF IEDLLFNKVT LADAGFIKQY GDCLGDIAAR DLICAQKFKG LTVLPPLLTD EMIAQYTSAL LAGTITSGW TFGAGAALQI PFAMQMAYRF NGIGVTQNVL YENQKLIANQ FNSAIGKIQD SLSSTASALG KLQDVVNHNA Q ALNTLVKQ LSSKFGAISS VLNDIFSRLD PPEAEVQIDR LITGRLQSLQ TYVTQQLIRA AEIRASANLA ATKMSECVLG QS KRVDFCG KGYHLMSFPQ SAPHGVVFLH VTYVPAQEKN FTTAPAICHD GKAHFPREGV FVSNGTHWFV TQRNFYEPQI ITT DNTFVS GNCDVVIGIV NNTVYDPLQP ELDSFKEELD KYFKNHTSPD VDLGDISGIN ASVVNIQKEI DRLNEVAKNL NESL IDLQE LGKYEQYIKD IRSLVPRGSP GSGYIPEAPR DGQAYVRKDG EWVLLSTFLG HHHHHH UniProtKB: Spike glycoprotein |
-Macromolecule #2: IY-2A Fab heavy chain
| Macromolecule | Name: IY-2A Fab heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.576566 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLVEWGAG LLKPSETLSL TCAVYGGSFS GYYWSWIRQP PGKGLEWIGL INHSGSTNYN PSLKSRVTIS LDTSKNQFSL KLTSVTAAD TAVYYCARGL GIFGVVTLSD VWGQGTTVTV SSASTKGPSV FPLAPSSKST SGGTAALGCL VKDYFPEPVT V SWNSGALT ...String: QVQLVEWGAG LLKPSETLSL TCAVYGGSFS GYYWSWIRQP PGKGLEWIGL INHSGSTNYN PSLKSRVTIS LDTSKNQFSL KLTSVTAAD TAVYYCARGL GIFGVVTLSD VWGQGTTVTV SSASTKGPSV FPLAPSSKST SGGTAALGCL VKDYFPEPVT V SWNSGALT SGVHTFPAVL QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKK VEPKSC |
-Macromolecule #3: IY-2A Fab light chain
| Macromolecule | Name: IY-2A Fab light chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.078279 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: NFMLTQPHSV SESPGKTVTI SCTGSSGSIA SNYVQWYQQR PGSAPTTVIY EDNQRPSGVP DRFSGSIDSS SNSASLTISG LRTEDEADY YCQSYDSGIW VFGGGTKLTV LGQPKAAPSV TLFPPSSEEL QANKATLVCL ISDFYPGAVT VAWKADSSPV K AGVETTTP ...String: NFMLTQPHSV SESPGKTVTI SCTGSSGSIA SNYVQWYQQR PGSAPTTVIY EDNQRPSGVP DRFSGSIDSS SNSASLTISG LRTEDEADY YCQSYDSGIW VFGGGTKLTV LGQPKAAPSV TLFPPSSEEL QANKATLVCL ISDFYPGAVT VAWKADSSPV K AGVETTTP SKQSNNKYAA SSYLSLTPEQ WKSHRSYSCQ VTHEGSTVEK TVAPTECS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 20mM Tris pH 7.5 150mM NaCl |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 258 K / Instrument: FEI VITROBOT MARK IV |
| Details | freshly mixed (3 min at room temperature) for complex formation |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 4685 / Average electron dose: 1.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8hhz: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)