[English] 日本語
 Yorodumi
Yorodumi- EMDB-34656: SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 3 YB13-... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 3 YB13-292 Fabs (3 RBD down) | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Spike protein / RBD / Antibody / Fab / Viral protein / VIRAL PROTEIN-IMMUNE SYSTEM complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / receptor ligand activity / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||||||||
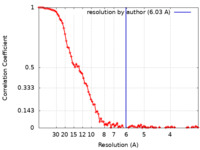
| Method | single particle reconstruction / cryo EM / Resolution: 6.03 Å | ||||||||||||
 Authors Authors | Liu B / Gao X / Chen Q / Li Z / Su M / He J / Xiong X | ||||||||||||
| Funding support | 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants. Authors: Haisheng Yu / Banghui Liu / Yudi Zhang / Xijie Gao / Qian Wang / Haitao Xiang / Xiaofang Peng / Caixia Xie / Yaping Wang / Peiyu Hu / Jingrong Shi / Quan Shi / Pingqian Zheng / Chengqian ...Authors: Haisheng Yu / Banghui Liu / Yudi Zhang / Xijie Gao / Qian Wang / Haitao Xiang / Xiaofang Peng / Caixia Xie / Yaping Wang / Peiyu Hu / Jingrong Shi / Quan Shi / Pingqian Zheng / Chengqian Feng / Guofang Tang / Xiaopan Liu / Liliangzi Guo / Xiumei Lin / Jiaojiao Li / Chuanyu Liu / Yaling Huang / Naibo Yang / Qiuluan Chen / Zimu Li / Mengzhen Su / Qihong Yan / Rongjuan Pei / Xinwen Chen / Longqi Liu / Fengyu Hu / Dan Liang / Bixia Ke / Changwen Ke / Feng Li / Jun He / Meiniang Wang / Ling Chen / Xiaoli Xiong / Xiaoping Tang /  Abstract: SARS-CoV-2 Omicron variants feature highly mutated spike proteins with extraordinary abilities in evading antibodies isolated earlier in the pandemic. Investigation of memory B cells from patients ...SARS-CoV-2 Omicron variants feature highly mutated spike proteins with extraordinary abilities in evading antibodies isolated earlier in the pandemic. Investigation of memory B cells from patients primarily with breakthrough infections with the Delta variant enables isolation of a number of neutralizing antibodies cross-reactive to heterologous variants of concern (VOCs) including Omicron variants (BA.1-BA.4). Structural studies identify altered complementarity determining region (CDR) amino acids and highly unusual heavy chain CDR2 insertions respectively in two representative cross-neutralizing antibodies-YB9-258 and YB13-292. These features are putatively introduced by somatic hypermutation and they are heavily involved in epitope recognition to broaden neutralization breadth. Previously, insertions/deletions were rarely reported for antiviral antibodies except for those induced by HIV-1 chronic infections. These data provide molecular mechanisms for cross-neutralization of heterologous SARS-CoV-2 variants by antibodies isolated from Delta variant infected patients with implications for future vaccination strategy. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34656.map.gz emd_34656.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34656-v30.xml emd-34656-v30.xml emd-34656.xml emd-34656.xml | 25.2 KB 25.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34656_fsc.xml emd_34656_fsc.xml | 11.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_34656.png emd_34656.png | 38.4 KB | ||
| Filedesc metadata |  emd-34656.cif.gz emd-34656.cif.gz | 7.8 KB | ||
| Others |  emd_34656_half_map_1.map.gz emd_34656_half_map_1.map.gz emd_34656_half_map_2.map.gz emd_34656_half_map_2.map.gz | 49.6 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34656 http://ftp.pdbj.org/pub/emdb/structures/EMD-34656 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34656 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34656 | HTTPS FTP |
-Validation report
| Summary document |  emd_34656_validation.pdf.gz emd_34656_validation.pdf.gz | 996.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34656_full_validation.pdf.gz emd_34656_full_validation.pdf.gz | 996.5 KB | Display | |
| Data in XML |  emd_34656_validation.xml.gz emd_34656_validation.xml.gz | 15.7 KB | Display | |
| Data in CIF |  emd_34656_validation.cif.gz emd_34656_validation.cif.gz | 21.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34656 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34656 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34656 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34656 | HTTPS FTP |
-Related structure data
| Related structure data |  8hc9MC  8hc2C  8hc3C  8hc4C  8hc5C  8hc6C  8hc7C  8hc8C  8hcaC  8hcbC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34656.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34656.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.65 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34656_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
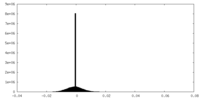
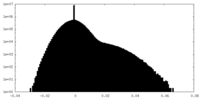
| Density Histograms |
-Half map: #1
| File | emd_34656_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
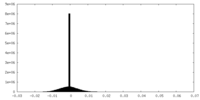
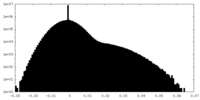
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 Omicron BA.1 spike trimer with three RBD down in compl...
| Entire | Name: SARS-CoV-2 Omicron BA.1 spike trimer with three RBD down in complex with three YB13-292 Fab |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 Omicron BA.1 spike trimer with three RBD down in compl...
| Supramolecule | Name: SARS-CoV-2 Omicron BA.1 spike trimer with three RBD down in complex with three YB13-292 Fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|
-Supramolecule #2: SARS-CoV-2 Omicron BA.1 spike protein
| Supramolecule | Name: SARS-CoV-2 Omicron BA.1 spike protein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: YB13-292 Fab
| Supramolecule | Name: YB13-292 Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 138.036188 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QCVNLTTRTQ LPPAYTNSFT RGVYYPDKVF RSSVLHSTQD LFLPFFSNVT WFHVISGTNG TKRFDNPVLP FNDGVYFASI EKSNIIRGW IFGTTLDSKT QSLLIVNNAT NVVIKVCEFQ FCNDPFLDHK NNKSWMESEF RVYSSANNCT FEYVSQPFLM D LEGKQGNF ...String: QCVNLTTRTQ LPPAYTNSFT RGVYYPDKVF RSSVLHSTQD LFLPFFSNVT WFHVISGTNG TKRFDNPVLP FNDGVYFASI EKSNIIRGW IFGTTLDSKT QSLLIVNNAT NVVIKVCEFQ FCNDPFLDHK NNKSWMESEF RVYSSANNCT FEYVSQPFLM D LEGKQGNF KNLREFVFKN IDGYFKIYSK HTPIIVREPE DLPQGFSALE PLVDLPIGIN ITRFQTLLAL HRSYLTPGDS SS GWTAGAA AYYVGYLQPR TFLLKYNENG TITDAVDCAL DPLSETKCTL KSFTVEKGIY QTSNFRVQPT ESIVRFPNIT NLC PFDEVF NATRFASVYA WNRKRISNCV ADYSVLYNLA PFFTFKCYGV SPTKLNDLCF TNVYADSFVI RGDEVRQIAP GQTG NIADY NYKLPDDFTG CVIAWNSNKL DSKVSGNYNY LYRLFRKSNL KPFERDISTE IYQAGNKPCN GVAGFNCYFP LRSYS FRPT YGVGHQPYRV VVLSFELLHA PATVCGPKKS TNLVKNKCVN FNFNGLKGTG VLTESNKKFL PFQQFGRDIA DTTDAV RDP QTLEILDITP CSFGGVSVIT PGTNTSNQVA VLYQGVNCTE VPVAIHADQL TPTWRVYSTG SNVFQTRAGC LIGAEYV NN SYECDIPIGA GICASYQTQT KSRSVASQSI IAYTMSLGAE NSVAYSNNSI AIPTNFTISV TTEILPVSMT KTSVDCTM Y ICGDSTECSN LLLQYGSFCT QLKRALTGIA VEQDKNTQEV FAQVKQIYKT PPIKYFGGFN FSQILPDPSK PSKRSPIED LLFNKVTLAD AGFIKQYGDC LGDIAARDLI CAQKFKGLTV LPPLLTDEMI AQYTSALLAG TITSGWTFGA GPALQIPFPM QMAYRFNGI GVTQNVLYEN QKLIANQFNS AIGKIQDSLS STPSALGKLQ DVVNHNAQAL NTLVKQLSSK FGAISSVLND I FSRLDPPE AEVQIDRLIT GRLQSLQTYV TQQLIRAAEI RASANLAATK MSECVLGQSK RVDFCGKGYH LMSFPQSAPH GV VFLHVTY VPAQEKNFTT APAICHDGKA HFPREGVFVS NGTHWFVTQR NFYEPQIITT DNTFVSGNCD VVIGIVNNTV YDP LQPELD SFKEELDKYF KNHTSPDVDL GDISGINASV VNIQKEIDRL NEVAKNLNES LIDLQELGKY EQYIKGSGRE NLYF QGGGG SGYIPEAPRD GQAYVRKDGE WVLLSTFLGH HHHHH UniProtKB: Spike glycoprotein |
-Macromolecule #2: Light chain of YB13-292 Fab
| Macromolecule | Name: Light chain of YB13-292 Fab / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.976648 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIVLTQSPLS LPVTPGEPAS ISCRSSQSLL RSNGYNYLDW YLQKPGQSPH LLIYLGSNRA SGVPDRFSGS GSGTDFTLKI SRVEAEDVG VYYCMQALQT PYTFGQGTNL EIKRTVAAPS VFIFPPSDEQ LKSGTASVVC LLNNFYPREA KVQWKVDNAL Q SGNSQESV ...String: DIVLTQSPLS LPVTPGEPAS ISCRSSQSLL RSNGYNYLDW YLQKPGQSPH LLIYLGSNRA SGVPDRFSGS GSGTDFTLKI SRVEAEDVG VYYCMQALQT PYTFGQGTNL EIKRTVAAPS VFIFPPSDEQ LKSGTASVVC LLNNFYPREA KVQWKVDNAL Q SGNSQESV TEQDSKDSTY SLSSTLTLSK ADYEKHKVYA CEVTHQGLSS PVTKSFNRGE C |
-Macromolecule #3: Heavy chain of YB13-292 Fab
| Macromolecule | Name: Heavy chain of YB13-292 Fab / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.283396 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGGG LVKPGGSLRL SCAASGFSFI TYNMNWVRQA PGKGLEWVSS ISSNILSSTS YIYYADSVKG RFTISRDDAA NSLFLQMNS LRVEDTAQYY CARTRSRSVR NCTSATCPVD AFDLWGQGTM VIVSSASTKG PSVFPLAPSS KSTSGGTAAL G CLVKDYFP ...String: EVQLVESGGG LVKPGGSLRL SCAASGFSFI TYNMNWVRQA PGKGLEWVSS ISSNILSSTS YIYYADSVKG RFTISRDDAA NSLFLQMNS LRVEDTAQYY CARTRSRSVR NCTSATCPVD AFDLWGQGTM VIVSSASTKG PSVFPLAPSS KSTSGGTAAL G CLVKDYFP EPVTVSWNSG ALTSGVHTFP AVLQSSGLYS LSSVVTVPSS SLGTQTYICN VNHKPSNTKV DKKVEPKSCD |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 30 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)