[English] 日本語
 Yorodumi
Yorodumi- EMDB-34652: SARS-CoV-2 wildtype S1 in complex with YB9-258 Fab and R1-32 Fab -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 wildtype S1 in complex with YB9-258 Fab and R1-32 Fab | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Spike protein / RBD / antibody / Fab / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / receptor ligand activity / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||||||||
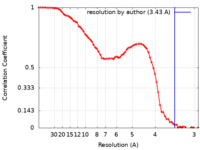
| Method | single particle reconstruction / cryo EM / Resolution: 3.43 Å | ||||||||||||
 Authors Authors | Liu B / Gao X / Chen Q / Li Z / Su M / He J / Xiong X | ||||||||||||
| Funding support | 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants. Authors: Haisheng Yu / Banghui Liu / Yudi Zhang / Xijie Gao / Qian Wang / Haitao Xiang / Xiaofang Peng / Caixia Xie / Yaping Wang / Peiyu Hu / Jingrong Shi / Quan Shi / Pingqian Zheng / Chengqian ...Authors: Haisheng Yu / Banghui Liu / Yudi Zhang / Xijie Gao / Qian Wang / Haitao Xiang / Xiaofang Peng / Caixia Xie / Yaping Wang / Peiyu Hu / Jingrong Shi / Quan Shi / Pingqian Zheng / Chengqian Feng / Guofang Tang / Xiaopan Liu / Liliangzi Guo / Xiumei Lin / Jiaojiao Li / Chuanyu Liu / Yaling Huang / Naibo Yang / Qiuluan Chen / Zimu Li / Mengzhen Su / Qihong Yan / Rongjuan Pei / Xinwen Chen / Longqi Liu / Fengyu Hu / Dan Liang / Bixia Ke / Changwen Ke / Feng Li / Jun He / Meiniang Wang / Ling Chen / Xiaoli Xiong / Xiaoping Tang /  Abstract: SARS-CoV-2 Omicron variants feature highly mutated spike proteins with extraordinary abilities in evading antibodies isolated earlier in the pandemic. Investigation of memory B cells from patients ...SARS-CoV-2 Omicron variants feature highly mutated spike proteins with extraordinary abilities in evading antibodies isolated earlier in the pandemic. Investigation of memory B cells from patients primarily with breakthrough infections with the Delta variant enables isolation of a number of neutralizing antibodies cross-reactive to heterologous variants of concern (VOCs) including Omicron variants (BA.1-BA.4). Structural studies identify altered complementarity determining region (CDR) amino acids and highly unusual heavy chain CDR2 insertions respectively in two representative cross-neutralizing antibodies-YB9-258 and YB13-292. These features are putatively introduced by somatic hypermutation and they are heavily involved in epitope recognition to broaden neutralization breadth. Previously, insertions/deletions were rarely reported for antiviral antibodies except for those induced by HIV-1 chronic infections. These data provide molecular mechanisms for cross-neutralization of heterologous SARS-CoV-2 variants by antibodies isolated from Delta variant infected patients with implications for future vaccination strategy. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34652.map.gz emd_34652.map.gz | 25.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34652-v30.xml emd-34652-v30.xml emd-34652.xml emd-34652.xml | 25.8 KB 25.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34652_fsc.xml emd_34652_fsc.xml | 8.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_34652.png emd_34652.png | 39.4 KB | ||
| Filedesc metadata |  emd-34652.cif.gz emd-34652.cif.gz | 7.5 KB | ||
| Others |  emd_34652_half_map_1.map.gz emd_34652_half_map_1.map.gz emd_34652_half_map_2.map.gz emd_34652_half_map_2.map.gz | 25.1 MB 25.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34652 http://ftp.pdbj.org/pub/emdb/structures/EMD-34652 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34652 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34652 | HTTPS FTP |
-Validation report
| Summary document |  emd_34652_validation.pdf.gz emd_34652_validation.pdf.gz | 774.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34652_full_validation.pdf.gz emd_34652_full_validation.pdf.gz | 774.2 KB | Display | |
| Data in XML |  emd_34652_validation.xml.gz emd_34652_validation.xml.gz | 12.4 KB | Display | |
| Data in CIF |  emd_34652_validation.cif.gz emd_34652_validation.cif.gz | 17.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34652 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34652 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34652 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34652 | HTTPS FTP |
-Related structure data
| Related structure data |  8hc5MC  8hc2C  8hc3C  8hc4C  8hc6C  8hc7C  8hc8C  8hc9C  8hcaC  8hcbC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34652.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34652.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.464 Å | ||||||||||||||||||||||||||||||||||||
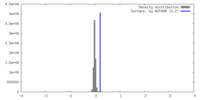
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34652_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
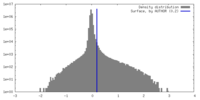
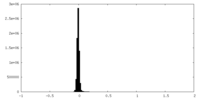
| Density Histograms |
-Half map: #1
| File | emd_34652_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
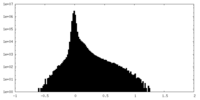
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 wildtype S1 in complex with YB9-258 Fab and R1-32 Fab
| Entire | Name: SARS-CoV-2 wildtype S1 in complex with YB9-258 Fab and R1-32 Fab |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 wildtype S1 in complex with YB9-258 Fab and R1-32 Fab
| Supramolecule | Name: SARS-CoV-2 wildtype S1 in complex with YB9-258 Fab and R1-32 Fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|
-Supramolecule #2: SARS-CoV_2 wildtype S1
| Supramolecule | Name: SARS-CoV_2 wildtype S1 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: YB9-258 Fab and R1-32 Fab
| Supramolecule | Name: YB9-258 Fab and R1-32 Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 75.688125 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF EYVSQPFLMD LEGKQGNFKN LREFVFKNID GYFKIYSKHT PINLVRDLPQ GFSALEPLVD LPIGINITRF QT LLALHRS YLTPGDSSSG WTAGAAAYYV GYLQPRTFLL KYNENGTITD AVDCALDPLS ETKCTLKSFT VEKGIYQTSN FRV QPTESI VRFPNITNLC PFGEVFNATR FASVYAWNRK RISNCVADYS VLYNSASFST FKCYGVSPTK LNDLCFTNVY ADSF VIRGD EVRQIAPGQT GKIADYNYKL PDDFTGCVIA WNSNNLDSKV GGNYNYLYRL FRKSNLKPFE RDISTEIYQA GSTPC NGVE GFNCYFPLQS YGFQPTNGVG YQPYRVVVLS FELLHAPATV CGPKKSTNLV KNKCVNFNFN GLTGTGVLTE SNKKFL PFQ QFGRDIADTT DAVRDPQTLE ILDITPCSFG GVSVITPGTN TSNQVAVLYQ DVNCTEVPVA IHADQLTPTW RVYSTGS NV FQTRAGCLIG AEHVNNSYEC DIPIGAGICA SYQT UniProtKB: Spike glycoprotein |
-Macromolecule #2: Heavy chain of YB9-258 Fab
| Macromolecule | Name: Heavy chain of YB9-258 Fab / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.126922 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGGG LIQPGGSLRL SCAASGLTVS SNYMHWVRQA PGKGLEWVSV LYAGGSAFYA DSVKGRFTIS RNNSKNTLYL QMNSLRAED TAIYYCARGL GDYLDSWGQG TLVTVSSAST KGPSVFPLAP SSKSTSGGTA ALGCLVKDYF PEPVTVSWNS G ALTSGVHT ...String: EVQLVESGGG LIQPGGSLRL SCAASGLTVS SNYMHWVRQA PGKGLEWVSV LYAGGSAFYA DSVKGRFTIS RNNSKNTLYL QMNSLRAED TAIYYCARGL GDYLDSWGQG TLVTVSSAST KGPSVFPLAP SSKSTSGGTA ALGCLVKDYF PEPVTVSWNS G ALTSGVHT FPAVLQSSGL YSLSSVVTVP SSSLGTQTYI CNVNHKPSNT KVDKKVEPKS CD |
-Macromolecule #3: Light chain of YB9-258
| Macromolecule | Name: Light chain of YB9-258 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.177754 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSS VSASVGDRVT ITCRASQGIG SWLAWYQQKP GKAPQLLIYA ASTLQSGVPP RFSGSGSGTD FTLTITSLQP EDFASYYCQ QANSVLALTF GGGTKVEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: DIQMTQSPSS VSASVGDRVT ITCRASQGIG SWLAWYQQKP GKAPQLLIYA ASTLQSGVPP RFSGSGSGTD FTLTITSLQP EDFASYYCQ QANSVLALTF GGGTKVEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #4: Heavy chain of R1-32 Fab
| Macromolecule | Name: Heavy chain of R1-32 Fab / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.840717 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGAE VKKPGSSVKV SCKASGGTFS SYAISWVRQA PGQGLEWMGG IIPILGIANY AQKFQGRVTI TADKSTSTAY MELSSLRSE DTAVYYCARE NGYSGYGAAA NFDLWGRGTL VTVSSASTKG PSVFPLAPSS KSTSGGTAAL GCLVKDYFPE P VTVSWNSG ...String: EVQLVESGAE VKKPGSSVKV SCKASGGTFS SYAISWVRQA PGQGLEWMGG IIPILGIANY AQKFQGRVTI TADKSTSTAY MELSSLRSE DTAVYYCARE NGYSGYGAAA NFDLWGRGTL VTVSSASTKG PSVFPLAPSS KSTSGGTAAL GCLVKDYFPE P VTVSWNSG ALTSGVHTFP AVLQSSGLYS LSSVVTVPSS SLGTQTYICN VNHKPSNTKV DKKVEPKSCD |
-Macromolecule #5: Light chain of R1-32 Fab
| Macromolecule | Name: Light chain of R1-32 Fab / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.382643 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSVLTQPPSV SGAPGQRVTI SCTGSSSNIG AGYDVHWYQQ LPGTAPKLLI YGNSNRPSGV PDRFSGSKSG TSASLAITGL QAEDEADYY CQSYDSSLSG SVFGGGTKLT VLGQPKAAPS VTLFPPSSEE LQANKATLVC LISDFYPGAV TVAWKADSSP V KAGVETTT ...String: QSVLTQPPSV SGAPGQRVTI SCTGSSSNIG AGYDVHWYQQ LPGTAPKLLI YGNSNRPSGV PDRFSGSKSG TSASLAITGL QAEDEADYY CQSYDSSLSG SVFGGGTKLT VLGQPKAAPS VTLFPPSSEE LQANKATLVC LISDFYPGAV TVAWKADSSP V KAGVETTT PSKQSNNKYA ASSYLSLTPE QWKSHRSYSC QVTHEGSTVE KTVAPT |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)