[English] 日本語
 Yorodumi
Yorodumi- EMDB-34597: Cryo-EM structure of the CBP catalytic core bound to the H4K12acK... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the CBP catalytic core bound to the H4K12acK16ac nucleosome, class 3 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Acetyl taransferase / Complex / Nucleosome / TRANSFERASE / TRANSFERASE-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationpeptide lactyltransferase (CoA-dependent) activity / NFE2L2 regulating ER-stress associated genes / NFE2L2 regulating inflammation associated genes / Activation of the TFAP2 (AP-2) family of transcription factors / histone H3K18 acetyltransferase activity / N-terminal peptidyl-lysine acetylation / LRR FLII-interacting protein 1 (LRRFIP1) activates type I IFN production / histone H3K27 acetyltransferase activity / NFE2L2 regulates pentose phosphate pathway genes / regulation of smoothened signaling pathway ...peptide lactyltransferase (CoA-dependent) activity / NFE2L2 regulating ER-stress associated genes / NFE2L2 regulating inflammation associated genes / Activation of the TFAP2 (AP-2) family of transcription factors / histone H3K18 acetyltransferase activity / N-terminal peptidyl-lysine acetylation / LRR FLII-interacting protein 1 (LRRFIP1) activates type I IFN production / histone H3K27 acetyltransferase activity / NFE2L2 regulates pentose phosphate pathway genes / regulation of smoothened signaling pathway / NFE2L2 regulating MDR associated enzymes / MRF binding / RUNX1 regulates transcription of genes involved in differentiation of myeloid cells / Regulation of gene expression in late stage (branching morphogenesis) pancreatic bud precursor cells / Regulation of FOXO transcriptional activity by acetylation / RUNX3 regulates NOTCH signaling / NOTCH4 Intracellular Domain Regulates Transcription / Regulation of NFE2L2 gene expression / Nuclear events mediated by NFE2L2 / Regulation of gene expression by Hypoxia-inducible Factor / negative regulation of transcription by RNA polymerase I / NOTCH3 Intracellular Domain Regulates Transcription / TRAF6 mediated IRF7 activation / NFE2L2 regulating tumorigenic genes / NFE2L2 regulating anti-oxidant/detoxification enzymes / embryonic digit morphogenesis / protein-lysine-acetyltransferase activity / protein acetylation / Notch-HLH transcription pathway / Formation of paraxial mesoderm / positive regulation of transforming growth factor beta receptor signaling pathway / acetyltransferase activity / homeostatic process / FOXO-mediated transcription of cell death genes / stimulatory C-type lectin receptor signaling pathway / Zygotic genome activation (ZGA) / TP53 Regulates Transcription of Genes Involved in Cytochrome C Release / histone acetyltransferase complex / canonical NF-kappaB signal transduction / negative regulation of tumor necrosis factor-mediated signaling pathway / Attenuation phase / cellular response to nutrient levels / histone acetyltransferase activity / histone acetyltransferase / positive regulation of double-strand break repair via homologous recombination / regulation of cellular response to heat / protein localization to CENP-A containing chromatin / Chromatin modifying enzymes / Replacement of protamines by nucleosomes in the male pronucleus / CENP-A containing nucleosome / : / Regulation of lipid metabolism by PPARalpha / NPAS4 regulates expression of target genes / Packaging Of Telomere Ends / Transferases; Acyltransferases; Transferring groups other than aminoacyl groups / Recognition and association of DNA glycosylase with site containing an affected purine / Cleavage of the damaged purine / Transcriptional and post-translational regulation of MITF-M expression and activity / CD209 (DC-SIGN) signaling / Deposition of new CENPA-containing nucleosomes at the centromere / BMAL1:CLOCK,NPAS2 activates circadian expression / SUMOylation of transcription cofactors / telomere organization / Recognition and association of DNA glycosylase with site containing an affected pyrimidine / Cleavage of the damaged pyrimidine / Interleukin-7 signaling / Activation of gene expression by SREBF (SREBP) / RNA Polymerase I Promoter Opening / epigenetic regulation of gene expression / Inhibition of DNA recombination at telomere / Assembly of the ORC complex at the origin of replication / Meiotic synapsis / Regulation of endogenous retroelements by the Human Silencing Hub (HUSH) complex / DNA methylation / Condensation of Prophase Chromosomes / Chromatin modifications during the maternal to zygotic transition (MZT) / SIRT1 negatively regulates rRNA expression / HCMV Late Events / ERCC6 (CSB) and EHMT2 (G9a) positively regulate rRNA expression / PRC2 methylates histones and DNA / Regulation of endogenous retroelements by KRAB-ZFP proteins / innate immune response in mucosa / Defective pyroptosis / HDACs deacetylate histones / Regulation of endogenous retroelements by Piwi-interacting RNAs (piRNAs) / RNA Polymerase I Promoter Escape / Nonhomologous End-Joining (NHEJ) / Heme signaling / lipopolysaccharide binding / Transcriptional regulation by small RNAs / Transcriptional activation of mitochondrial biogenesis / PPARA activates gene expression / Formation of the beta-catenin:TCF transactivating complex / Cytoprotection by HMOX1 / Activated PKN1 stimulates transcription of AR (androgen receptor) regulated genes KLK2 and KLK3 / RUNX1 regulates genes involved in megakaryocyte differentiation and platelet function / HDMs demethylate histones / G2/M DNA damage checkpoint / NoRC negatively regulates rRNA expression / chromatin DNA binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
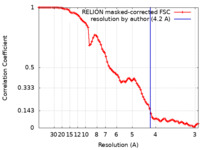
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Kikuchi M / Morita S / Wakamori M / Shin S / Uchikubo-Kamo T / Shirouzu M / Umehara T | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Epigenetic mechanisms to propagate histone acetylation by p300/CBP. Authors: Masaki Kikuchi / Satoshi Morita / Masatoshi Wakamori / Shin Sato / Tomomi Uchikubo-Kamo / Takehiro Suzuki / Naoshi Dohmae / Mikako Shirouzu / Takashi Umehara /  Abstract: Histone acetylation is important for the activation of gene transcription but little is known about its direct read/write mechanisms. Here, we report cryogenic electron microscopy structures in which ...Histone acetylation is important for the activation of gene transcription but little is known about its direct read/write mechanisms. Here, we report cryogenic electron microscopy structures in which a p300/CREB-binding protein (CBP) multidomain monomer recognizes histone H4 N-terminal tail (NT) acetylation (ac) in a nucleosome and acetylates non-H4 histone NTs within the same nucleosome. p300/CBP not only recognized H4NTac via the bromodomain pocket responsible for reading, but also interacted with the DNA minor grooves via the outside of that pocket. This directed the catalytic center of p300/CBP to one of the non-H4 histone NTs. The primary target that p300 writes by reading H4NTac was H2BNT, and H2BNTac promoted H2A-H2B dissociation from the nucleosome. We propose a model in which p300/CBP replicates histone N-terminal tail acetylation within the H3-H4 tetramer to inherit epigenetic storage, and transcribes it from the H3-H4 tetramer to the H2B-H2A dimers to activate context-dependent gene transcription through local nucleosome destabilization. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34597.map.gz emd_34597.map.gz | 54.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34597-v30.xml emd-34597-v30.xml emd-34597.xml emd-34597.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34597_fsc.xml emd_34597_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_34597.png emd_34597.png | 72.4 KB | ||
| Masks |  emd_34597_msk_1.map emd_34597_msk_1.map | 59.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-34597.cif.gz emd-34597.cif.gz | 6.7 KB | ||
| Others |  emd_34597_half_map_1.map.gz emd_34597_half_map_1.map.gz emd_34597_half_map_2.map.gz emd_34597_half_map_2.map.gz | 46 MB 46 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34597 http://ftp.pdbj.org/pub/emdb/structures/EMD-34597 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34597 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34597 | HTTPS FTP |
-Related structure data
| Related structure data |  8hanMC  8hagC  8hahC  8haiC  8hajC  8hakC  8halC  8hamC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34597.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34597.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.47 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_34597_msk_1.map emd_34597_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_34597_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_34597_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : the CBP catalytic core bound to the H4K12acK16ac nucleosome
| Entire | Name: the CBP catalytic core bound to the H4K12acK16ac nucleosome |
|---|---|
| Components |
|
-Supramolecule #1: the CBP catalytic core bound to the H4K12acK16ac nucleosome
| Supramolecule | Name: the CBP catalytic core bound to the H4K12acK16ac nucleosome type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: CBP was expressed in insect cells. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Histone H3.1
| Macromolecule | Name: Histone H3.1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 15.305969 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ARTKQTARKS TGGKAPRKQL ATKAARKSAP ATGGVKKPHR YRPGTVALRE IRRYQKSTEL LIRKLPFQRL VREIAQDFKT DLRFQSSAV MALQEACEAY LVGLFEDTNL CAIHAKRVTI MPKDIQLARR IRGERA UniProtKB: Histone H3.1 |
-Macromolecule #2: Histone H4
| Macromolecule | Name: Histone H4 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.345289 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SGRGKGGKGL G(ALY)GGA(ALY)RHRK VLRDNIQGIT KPAIRRLARR GGVKRISGLI YEETRGVLKV FLENVIRDAV TY TEHAKRK TVTAMDVVYA LKRQGRTLYG FGG |
-Macromolecule #3: Histone H2A type 1-B/E
| Macromolecule | Name: Histone H2A type 1-B/E / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.034355 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SGRGKQGGKA RAKAKTRSSR AGLQFPVGRV HRLLRKGNYS ERVGAGAPVY LAAVLEYLTA EILELAGNAA RDNKKTRIIP RHLQLAIRN DEELNKLLGR VTIAQGGVLP NIQAVLLPKK TESHHKAKGK UniProtKB: Histone H2A type 1-B/E |
-Macromolecule #4: Histone H2B type 1-J
| Macromolecule | Name: Histone H2B type 1-J / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.804045 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: PEPAKSAPAP KKGSKKAVTK AQKKDGKKRK RSRKESYSIY VYKVLKQVHP DTGISSKAMG IMNSFVNDIF ERIAGEASRL AHYNKRSTI TSREIQTAVR LLLPGELAKH AVSEGTKAVT KYTSAK UniProtKB: Histone H2B type 1-J |
-Macromolecule #5: CREB-binding protein
| Macromolecule | Name: CREB-binding protein / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO / EC number: histone acetyltransferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 92.690906 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GSSGSSGIFK PEELRQALMP TLEALYRQDP ESLPFRQPVD PQLLGIPDYF DIVKNPMDLS TIKRKLDTGQ YQEPWQYVDD VWLMFNNAW LYNRKTSRVY KFCSKLAEVF EQEIDPVMQS LGYCCGRKYE FSPQTLCCYG KQLCTIPRDA AYYSYQNRYH F CEKCFTEI ...String: GSSGSSGIFK PEELRQALMP TLEALYRQDP ESLPFRQPVD PQLLGIPDYF DIVKNPMDLS TIKRKLDTGQ YQEPWQYVDD VWLMFNNAW LYNRKTSRVY KFCSKLAEVF EQEIDPVMQS LGYCCGRKYE FSPQTLCCYG KQLCTIPRDA AYYSYQNRYH F CEKCFTEI QGENVTLGDD PSQPQTTISK DQFEKKKNDT LDPEPFVDCK ECGRKMHQIC VLHYDIIWPS GFVCDNCLKK TG RPRKENK FSAKRLQTTR LGNHLEDRVN KFLRRQNHPE AGEVFVRVVA SSDKTVEVKP GMKSRFVDSG EMSESFPYRT KAL FAFEEI DGVDVCFFGM HVQEYGSDCP PPNTRRVYIS YLDSIHFFRP RCLRTAVYHE ILIGYLEYVK KLGYVTGHIW ACPP SEGDD YIFHCHPPDQ KIPKPKRLQE WYKKMLDKAF AERIIHDYKD IFKQATEDRL TSAKELPYFE GDFWPNVLEE SIKEL EQEE EERKKEESTA ASETTEGSQG DSKNAKKKNN KKTNKNKSSI SRANKKKPSM PNVSNDLSQK LYATMEKHKE VFFVIH LHA GPVINTLPPI VDPDPLLSCD LMDGRDAFLT LARDKHWEFS SLRRSKWSTL CMLVELHTQG QDRFVYTCNE CKHHVET RW HCTVCEDYDL CINCYNTKSH AHKMVKWGLG LDDEGSSQGE PQSKSPQESR RLSIQRCIQS LVHACQCRNA NCSLPSCQ K MKRVVQHTKG CKRKTNGGCP VCKQLIALCC YHAKHCQENK CPVPFCLNIK HKLRQQQIQH RLQQAQLMRR RMATMN UniProtKB: CREB-binding protein |
-Macromolecule #6: DNA (180-mer)
| Macromolecule | Name: DNA (180-mer) / type: dna / ID: 6 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 55.560527 KDa |
| Sequence | String: (DA)(DT)(DC)(DC)(DG)(DT)(DC)(DC)(DG)(DT) (DT)(DA)(DC)(DC)(DG)(DC)(DC)(DA)(DT)(DC) (DA)(DA)(DT)(DA)(DT)(DC)(DC)(DA)(DC) (DC)(DT)(DG)(DC)(DA)(DG)(DA)(DT)(DT)(DC) (DT) (DA)(DC)(DC)(DA)(DA)(DA) ...String: (DA)(DT)(DC)(DC)(DG)(DT)(DC)(DC)(DG)(DT) (DT)(DA)(DC)(DC)(DG)(DC)(DC)(DA)(DT)(DC) (DA)(DA)(DT)(DA)(DT)(DC)(DC)(DA)(DC) (DC)(DT)(DG)(DC)(DA)(DG)(DA)(DT)(DT)(DC) (DT) (DA)(DC)(DC)(DA)(DA)(DA)(DA)(DG) (DT)(DG)(DT)(DA)(DT)(DT)(DT)(DG)(DG)(DA) (DA)(DA) (DC)(DT)(DG)(DC)(DT)(DC)(DC) (DA)(DT)(DC)(DA)(DA)(DA)(DA)(DG)(DG)(DC) (DA)(DT)(DG) (DT)(DT)(DC)(DA)(DG)(DC) (DT)(DG)(DA)(DA)(DT)(DT)(DC)(DA)(DG)(DC) (DT)(DG)(DA)(DA) (DC)(DA)(DT)(DG)(DC) (DC)(DT)(DT)(DT)(DT)(DG)(DA)(DT)(DG)(DG) (DA)(DG)(DC)(DA)(DG) (DT)(DT)(DT)(DC) (DC)(DA)(DA)(DA)(DT)(DA)(DC)(DA)(DC)(DT) (DT)(DT)(DT)(DG)(DG)(DT) (DA)(DG)(DA) (DA)(DT)(DC)(DT)(DG)(DC)(DA)(DG)(DG)(DT) (DG)(DG)(DA)(DT)(DA)(DT)(DT) (DG)(DA) (DT)(DG)(DG)(DC)(DG)(DG)(DT)(DA)(DA)(DC) (DG)(DG)(DA)(DC)(DG)(DG)(DA)(DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.7 µm / Nominal defocus min: 0.9 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)