+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 BA.2.75 spike glycoprotein (closed state 2) | |||||||||||||||||||||
 Map data Map data | 0.0735 | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||
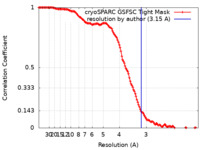
| Method | single particle reconstruction / cryo EM / Resolution: 3.15 Å | |||||||||||||||||||||
 Authors Authors | Anraku Y / Tabata-Sasaki K / Kita S / Fukuhara H / Maenaka K / Hashiguchi T | |||||||||||||||||||||
| Funding support |  Japan, 6 items Japan, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Cell Host Microbe / Year: 2022 Journal: Cell Host Microbe / Year: 2022Title: Virological characteristics of the SARS-CoV-2 Omicron BA.2.75 variant. Authors: Akatsuki Saito / Tomokazu Tamura / Jiri Zahradnik / Sayaka Deguchi / Koshiro Tabata / Yuki Anraku / Izumi Kimura / Jumpei Ito / Daichi Yamasoba / Hesham Nasser / Mako Toyoda / Kayoko Nagata ...Authors: Akatsuki Saito / Tomokazu Tamura / Jiri Zahradnik / Sayaka Deguchi / Koshiro Tabata / Yuki Anraku / Izumi Kimura / Jumpei Ito / Daichi Yamasoba / Hesham Nasser / Mako Toyoda / Kayoko Nagata / Keiya Uriu / Yusuke Kosugi / Shigeru Fujita / Maya Shofa / Mst Monira Begum / Ryo Shimizu / Yoshitaka Oda / Rigel Suzuki / Hayato Ito / Naganori Nao / Lei Wang / Masumi Tsuda / Kumiko Yoshimatsu / Jin Kuramochi / Shunsuke Kita / Kaori Sasaki-Tabata / Hideo Fukuhara / Katsumi Maenaka / Yuki Yamamoto / Tetsuharu Nagamoto / Hiroyuki Asakura / Mami Nagashima / Kenji Sadamasu / Kazuhisa Yoshimura / Takamasa Ueno / Gideon Schreiber / Akifumi Takaori-Kondo / / Kotaro Shirakawa / Hirofumi Sawa / Takashi Irie / Takao Hashiguchi / Kazuo Takayama / Keita Matsuno / Shinya Tanaka / Terumasa Ikeda / Takasuke Fukuhara / Kei Sato /    Abstract: The SARS-CoV-2 Omicron BA.2.75 variant emerged in May 2022. BA.2.75 is a BA.2 descendant but is phylogenetically distinct from BA.5, the currently predominant BA.2 descendant. Here, we show that BA.2. ...The SARS-CoV-2 Omicron BA.2.75 variant emerged in May 2022. BA.2.75 is a BA.2 descendant but is phylogenetically distinct from BA.5, the currently predominant BA.2 descendant. Here, we show that BA.2.75 has a greater effective reproduction number and different immunogenicity profile than BA.5. We determined the sensitivity of BA.2.75 to vaccinee and convalescent sera as well as a panel of clinically available antiviral drugs and antibodies. Antiviral drugs largely retained potency, but antibody sensitivity varied depending on several key BA.2.75-specific substitutions. The BA.2.75 spike exhibited a profoundly higher affinity for its human receptor, ACE2. Additionally, the fusogenicity, growth efficiency in human alveolar epithelial cells, and intrinsic pathogenicity in hamsters of BA.2.75 were greater than those of BA.2. Our multilevel investigations suggest that BA.2.75 acquired virological properties independent of BA.5, and the potential risk of BA.2.75 to global health is greater than that of BA.5. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34222.map.gz emd_34222.map.gz | 108.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34222-v30.xml emd-34222-v30.xml emd-34222.xml emd-34222.xml | 21.2 KB 21.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34222_fsc.xml emd_34222_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_34222.png emd_34222.png | 84.4 KB | ||
| Masks |  emd_34222_msk_1.map emd_34222_msk_1.map | 216 MB |  Mask map Mask map | |
| Others |  emd_34222_half_map_1.map.gz emd_34222_half_map_1.map.gz emd_34222_half_map_2.map.gz emd_34222_half_map_2.map.gz | 200.4 MB 200.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34222 http://ftp.pdbj.org/pub/emdb/structures/EMD-34222 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34222 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34222 | HTTPS FTP |
-Validation report
| Summary document |  emd_34222_validation.pdf.gz emd_34222_validation.pdf.gz | 890.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34222_full_validation.pdf.gz emd_34222_full_validation.pdf.gz | 890.2 KB | Display | |
| Data in XML |  emd_34222_validation.xml.gz emd_34222_validation.xml.gz | 21.6 KB | Display | |
| Data in CIF |  emd_34222_validation.cif.gz emd_34222_validation.cif.gz | 28 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34222 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34222 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34222 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34222 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34222.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34222.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 0.0735 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.005 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_34222_msk_1.map emd_34222_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: 0.219
| File | emd_34222_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 0.219 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: 0.219
| File | emd_34222_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 0.219 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-COV-2 BA.2.75 spike glycoprotein
| Entire | Name: SARS-COV-2 BA.2.75 spike glycoprotein |
|---|---|
| Components |
|
-Supramolecule #1: SARS-COV-2 BA.2.75 spike glycoprotein
| Supramolecule | Name: SARS-COV-2 BA.2.75 spike glycoprotein / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 420 KDa |
-Macromolecule #1: SARS-CoV-2 BA.2.75 spike glycoprotein
| Macromolecule | Name: SARS-CoV-2 BA.2.75 spike glycoprotein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: LLMGCVAETG SSQCVNLITR TQSYTNSFTR GVYYPDKVFR SSVLHSTQDL FLPFFSNVTW FHAIHVSGTN GTKRFDNPVL PFNDGVYFAS TEKSNIIRGW IFGTTLDSKT QSLLIVNNAT NVVIKVCEFQ FCNDPFLDVY YHENNKSRME SELRVYSSAN NCTFEYVSQP ...String: LLMGCVAETG SSQCVNLITR TQSYTNSFTR GVYYPDKVFR SSVLHSTQDL FLPFFSNVTW FHAIHVSGTN GTKRFDNPVL PFNDGVYFAS TEKSNIIRGW IFGTTLDSKT QSLLIVNNAT NVVIKVCEFQ FCNDPFLDVY YHENNKSRME SELRVYSSAN NCTFEYVSQP FLMDLEGKQG NFKNLREFVF KNIDGYFKIY SKHTPVNLGR DLPQGFSALE PLVDLPIGIN ITRFQTLLAL HRSYLTPGDS SSSWTAGAAA YYVGYLQPRT FLLKYNENGT ITDAVDCALD PLSETKCTLK SFTVEKGIYQ TSNFRVQPTE SIVRFPNITN LCPFHEVFNA TRFASVYAWN RKRISNCVAD YSVLYNFAPF FAFKCYGVSP TKLNDLCFTN VYADSFVIRG NEVSQIAPGQ TGNIADYNYK LPDDFTGCVI AWNSNKLDSK VSGNYNYLYR LFRKSKLKPF ERDISTEIYQ AGNKPCNGVA GFNCYFPLQS YGFRPTYGVG HQPYRVVVLS FELLHAPATV CGPKKSTNLV KNKCVNFNFN GLTGTGVLTE SNKKFLPFQQ FGRDIADTTD AVRDPQTLEI LDITPCSFGG VSVITPGTNT SNQVAVLYQG VNCTEVPVAI HADQLTPTWR VYSTGSNVFQ TRAGCLIGAE YVNNSYECDI PIGAGICASY QTQTKSHGSA GSVASQSIIA YTMSLGAENS VAYSNNSIAI PTNFTISVTT EILPVSMTKT SVDCTMYICG DSTECSNLLL QYGSFCTQLK RALTGIAVEQ DKNTQEVFAQ VKQIYKTPPI KYFGGFNFSQ ILSDPSKPSK RSPIEDLLFN KVTLADAGFI KQYGDCLGDI AARDLICAQK FKGLTVLPPL LTDEMIAQYT SALLAGTITS GWTFGAGPAL QIPFPMQMAY RFNGIGVTQN VLYENQKLIA NQFNSAIGKI QDSLSSTPSA LGKLQDVVNH NAQALNTLVK QLSSKFGAIS SVLNDIFSRL DPPEAEVQID RLITGRLQSL QTYVTQQLIR AAEIRASANL AATKMSECVL GQSKRVDFCG KGYHLMSFPQ SAPHGVVFLH VTYVPAQEKN FTTAPAICHD GKAHFPREGV FVSNGTHWFV TQRNFYEPQI ITTDNTFVSG NCDVVIGIVN NTVYDPLQPE LDSFKEELDK YFKNHTSPDV DLGDISGINA SVVNIQKEID RLNEVAKNLN ESLIDLQELG KYEQYIASSG YIPEAPRDGQ AYVRKDGEWV LLSTFLEGTK HHHHHH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL |
|---|---|
| Buffer | pH: 7.4 Details: Octyl glucoside solution was added to PBS solution to a final concentration of 0.01% |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 291 K / Instrument: FEI VITROBOT MARK IV / Details: blotting time 5 s and blotting force 5.. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number real images: 3308 / Average exposure time: 1.5 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)