[English] 日本語
 Yorodumi
Yorodumi- EMDB-3405: Mammalian 80S Ribosomes Associate with a Novel Vesicular Organelle -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3405 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mammalian 80S Ribosomes Associate with a Novel Vesicular Organelle | |||||||||
 Map data Map data | Reconstruction of Ribosome Associated Vesicle membrane-bound 80S ribosome | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ribosome / secretion / vesicle | |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM | |||||||||
 Authors Authors | Carter SD / Hampton CM / Langlois R / Grassucci RA / Farino ZJ / Morgenstern TM / Rice WJ / Velasco KR / Wigge C / Xu Y ...Carter SD / Hampton CM / Langlois R / Grassucci RA / Farino ZJ / Morgenstern TM / Rice WJ / Velasco KR / Wigge C / Xu Y / Koller A / Melero R / Mitchell WG / Yi E / Aguilar JI / Levy ES / Greenberg NL / Li W / Courel M / Mahata SK / Freyberg R / Javitch JA / Di Paolo G / Chen EI / Chan RB / Carazo JM / Area-Gomez E / Jensen GJ / Frank J / Freyberg Z | |||||||||
 Citation Citation |  Journal: NAT.COMMUN. Journal: NAT.COMMUN.Title: Mammalian 80S Ribosomes Associate with a Novel Vesicular Organelle Authors: Carter SD / Hampton CM / Langlois R / Grassucci RA / Farino ZJ / Morgenstern TM / Rice WJ / Velasco KR / Wigge C / Xu Y / Koller A / Melero R / Mitchell WG / Yi E / Aguilar JI / Levy ES / ...Authors: Carter SD / Hampton CM / Langlois R / Grassucci RA / Farino ZJ / Morgenstern TM / Rice WJ / Velasco KR / Wigge C / Xu Y / Koller A / Melero R / Mitchell WG / Yi E / Aguilar JI / Levy ES / Greenberg NL / Li W / Courel M / Mahata SK / Freyberg R / Javitch JA / Di Paolo G / Chen EI / Chan RB / Carazo JM / Area-Gomez E / Jensen GJ / Frank J / Freyberg Z | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3405.map.gz emd_3405.map.gz | 22.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3405-v30.xml emd-3405-v30.xml emd-3405.xml emd-3405.xml | 10.7 KB 10.7 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-3406.tif EMD-3406.tif | 664.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3405 http://ftp.pdbj.org/pub/emdb/structures/EMD-3405 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3405 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3405 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3405.map.gz / Format: CCP4 / Size: 39.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3405.map.gz / Format: CCP4 / Size: 39.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of Ribosome Associated Vesicle membrane-bound 80S ribosome | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.62 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
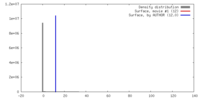
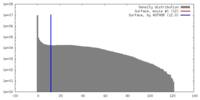
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Ribosome Associated Vesicle membrane-bound 80S ribosome
| Entire | Name: Ribosome Associated Vesicle membrane-bound 80S ribosome |
|---|---|
| Components |
|
-Supramolecule #1000: Ribosome Associated Vesicle membrane-bound 80S ribosome
| Supramolecule | Name: Ribosome Associated Vesicle membrane-bound 80S ribosome type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Supramolecule #1: Membrane-bound 80S ribosome
| Supramolecule | Name: Membrane-bound 80S ribosome / type: complex / ID: 1 / Name.synonym: 80S ribosome / Recombinant expression: No / Ribosome-details: ribosome-eukaryote: ALL |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 3 MDa / Theoretical: 3 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Grid | Details: 200 mesh gold grid with thin carbon support |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 150 K / Instrument: FEI VITROBOT MARK IV / Method: Blot for 8 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Min: 80 K / Max: 93 K / Average: 80 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 100,000 times magnification |
| Details | Weak beam illumination |
| Date | Mar 30, 2014 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 100 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 18977 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.26 mm / Nominal defocus max: 6.0 µm / Nominal defocus min: 6.0 µm / Nominal magnification: 15500 |
| Sample stage | Specimen holder model: GATAN HELIUM / Tilt series - Axis1 - Min angle: -60 ° / Tilt series - Axis1 - Max angle: 60 ° |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | The subtomograms were manually selected using the EMAN2 software package. |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution method: OTHER / Software - Name: ETOMO/Serial, EM Details: Final maps were calculated from five averaged datasets Number subtomograms used: 1230 |
| Final angle assignment | Details: SPIDER: theta +/- 60 degrees |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)