[English] 日本語
 Yorodumi
Yorodumi- EMDB-3390: Cryo-EM structure of a human APC/C mutant with Apc1 300s loop del... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3390 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a human APC/C mutant with Apc1 300s loop deleted in complex with TAME | |||||||||
 Map data Map data | Single particle reconstruction of a human APC/C mutant with Apc1 300s loop deleted in complex with TAME | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cell cycle / phosphorylation / mitosis / ubiquitination | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of synapse maturation / Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase / regulation of mitotic cell cycle spindle assembly checkpoint / Inactivation of APC/C via direct inhibition of the APC/C complex / APC/C:Cdc20 mediated degradation of mitotic proteins / positive regulation of synaptic plasticity / anaphase-promoting complex / regulation of meiotic cell cycle / Aberrant regulation of mitotic exit in cancer due to RB1 defects / anaphase-promoting complex-dependent catabolic process ...positive regulation of synapse maturation / Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase / regulation of mitotic cell cycle spindle assembly checkpoint / Inactivation of APC/C via direct inhibition of the APC/C complex / APC/C:Cdc20 mediated degradation of mitotic proteins / positive regulation of synaptic plasticity / anaphase-promoting complex / regulation of meiotic cell cycle / Aberrant regulation of mitotic exit in cancer due to RB1 defects / anaphase-promoting complex-dependent catabolic process / protein branched polyubiquitination / metaphase/anaphase transition of mitotic cell cycle / Phosphorylation of the APC/C / regulation of exit from mitosis / positive regulation of mitotic metaphase/anaphase transition / positive regulation of dendrite morphogenesis / protein K11-linked ubiquitination / regulation of mitotic metaphase/anaphase transition / ubiquitin-ubiquitin ligase activity / mitotic metaphase chromosome alignment / Regulation of APC/C activators between G1/S and early anaphase / Transcriptional Regulation by VENTX / cullin family protein binding / enzyme-substrate adaptor activity / positive regulation of axon extension / protein K48-linked ubiquitination / intercellular bridge / heterochromatin / nuclear periphery / APC/C:Cdc20 mediated degradation of Cyclin B / regulation of mitotic cell cycle / APC-Cdc20 mediated degradation of Nek2A / Autodegradation of Cdh1 by Cdh1:APC/C / APC/C:Cdc20 mediated degradation of Securin / Assembly of the pre-replicative complex / Cdc20:Phospho-APC/C mediated degradation of Cyclin A / APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 / G protein-coupled receptor binding / brain development / kinetochore / CDK-mediated phosphorylation and removal of Cdc6 / spindle / neuron projection development / ubiquitin-protein transferase activity / mitotic spindle / Separation of Sister Chromatids / ubiquitin protein ligase activity / nervous system development / mitotic cell cycle / Antigen processing: Ubiquitination & Proteasome degradation / microtubule cytoskeleton / Senescence-Associated Secretory Phenotype (SASP) / protein phosphatase binding / ubiquitin-dependent protein catabolic process / molecular adaptor activity / cell differentiation / protein ubiquitination / negative regulation of gene expression / cell division / ubiquitin protein ligase binding / centrosome / nucleolus / zinc ion binding / nucleoplasm / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / unidentified (others) Homo sapiens (human) / unidentified (others) | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 4.0 Å | |||||||||
 Authors Authors | Zhang S / Chang L / Alfieri C / Zhang Z / Yang J / Maslen S / Skehel M / Barford D | |||||||||
 Citation Citation |  Journal: Nature / Year: 2016 Journal: Nature / Year: 2016Title: Molecular mechanism of APC/C activation by mitotic phosphorylation. Authors: Suyang Zhang / Leifu Chang / Claudio Alfieri / Ziguo Zhang / Jing Yang / Sarah Maslen / Mark Skehel / David Barford /  Abstract: In eukaryotes, the anaphase-promoting complex (APC/C, also known as the cyclosome) regulates the ubiquitin-dependent proteolysis of specific cell-cycle proteins to coordinate chromosome segregation ...In eukaryotes, the anaphase-promoting complex (APC/C, also known as the cyclosome) regulates the ubiquitin-dependent proteolysis of specific cell-cycle proteins to coordinate chromosome segregation in mitosis and entry into the G1 phase. The catalytic activity of the APC/C and its ability to specify the destruction of particular proteins at different phases of the cell cycle are controlled by its interaction with two structurally related coactivator subunits, Cdc20 and Cdh1. Coactivators recognize substrate degrons, and enhance the affinity of the APC/C for its cognate E2 (refs 4-6). During mitosis, cyclin-dependent kinase (Cdk) and polo-like kinase (Plk) control Cdc20- and Cdh1-mediated activation of the APC/C. Hyperphosphorylation of APC/C subunits, notably Apc1 and Apc3, is required for Cdc20 to activate the APC/C, whereas phosphorylation of Cdh1 prevents its association with the APC/C. Since both coactivators associate with the APC/C through their common C-box and Ile-Arg tail motifs, the mechanism underlying this differential regulation is unclear, as is the role of specific APC/C phosphorylation sites. Here, using cryo-electron microscopy and biochemical analysis, we define the molecular basis of how phosphorylation of human APC/C allows for its control by Cdc20. An auto-inhibitory segment of Apc1 acts as a molecular switch that in apo unphosphorylated APC/C interacts with the C-box binding site and obstructs engagement of Cdc20. Phosphorylation of the auto-inhibitory segment displaces it from the C-box-binding site. Efficient phosphorylation of the auto-inhibitory segment, and thus relief of auto-inhibition, requires the recruitment of Cdk-cyclin in complex with a Cdk regulatory subunit (Cks) to a hyperphosphorylated loop of Apc3. We also find that the small-molecule inhibitor, tosyl-l-arginine methyl ester, preferentially suppresses APC/C(Cdc20) rather than APC/C(Cdh1), and interacts with the binding sites of both the C-box and Ile-Arg tail motifs. Our results reveal the mechanism for the regulation of mitotic APC/C by phosphorylation and provide a rationale for the development of selective inhibitors of this state. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3390.map.gz emd_3390.map.gz | 9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3390-v30.xml emd-3390-v30.xml emd-3390.xml emd-3390.xml | 24.3 KB 24.3 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-3390.jpg EMD-3390.jpg | 79.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3390 http://ftp.pdbj.org/pub/emdb/structures/EMD-3390 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3390 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3390 | HTTPS FTP |
-Related structure data
| Related structure data |  3385C  3386C  3387C  3388C  3389C  5g04C  5g05C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3390.map.gz / Format: CCP4 / Size: 68.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3390.map.gz / Format: CCP4 / Size: 68.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Single particle reconstruction of a human APC/C mutant with Apc1 300s loop deleted in complex with TAME | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
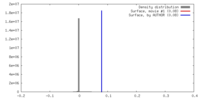
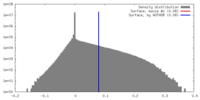
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Recombinant human APC/C mutant with Apc1 300s loop deleted in com...
+Supramolecule #1000: Recombinant human APC/C mutant with Apc1 300s loop deleted in com...
+Macromolecule #1: Anaphase-promoting complex subunit 1
+Macromolecule #2: Anaphase-promoting complex subunit 2
+Macromolecule #3: Anaphase-promoting complex subunit 3
+Macromolecule #4: Anaphase-promoting complex subunit 4
+Macromolecule #5: Anaphase-promoting complex subunit 5
+Macromolecule #6: Anaphase-promoting complex subunit 6
+Macromolecule #7: Anaphase-promoting complex subunit 7
+Macromolecule #8: Anaphase-promoting complex subunit 8
+Macromolecule #9: Anaphase-promoting complex subunit 10
+Macromolecule #10: Anaphase-promoting complex subunit 11
+Macromolecule #11: Anaphase-promoting complex subunit 12
+Macromolecule #12: Anaphase-promoting complex subunit 13
+Macromolecule #13: Anaphase-promoting complex subunit 15
+Macromolecule #14: Anaphase-promoting complex subunit 16
+Macromolecule #15: tosyl-L-arginine methyl ester
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.15 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 20mM Hepes, 150mM NaCl, 0.005mM TCEP |
| Staining | Type: NEGATIVE / Details: Vitrification in liquid ethane |
| Grid | Details: 200 mesh Quantifoil R2/2 copper grid with thin carbon support, treated with a 9:1 argon:oxygen plasma cleaner for 20-40s |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 100 K / Instrument: FEI VITROBOT MARK III Method: The grids were incubated for 30s at 4 degree and 100% humidity before blotting for 5s and plunging into liquid ethane |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Min: 90 K / Max: 120 K / Average: 105 K |
| Specialist optics | Energy filter - Name: FEI |
| Date | Dec 5, 2015 |
| Image recording | Category: CCD / Film or detector model: FEI FALCON II (4k x 4k) / Number real images: 1500 / Average electron dose: 27 e/Å2 Details: The exposure time for each micrograph was 2s at a dose rate of 27 electrons/angstrom2/s. 34 movie frames were recorded for each micrograph. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 78000 |
| Sample stage | Specimen holder: liquid nitrogen cooled / Specimen holder model: OTHER |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Particles were selected by automatic particle picking in RELION |
|---|---|
| CTF correction | Details: Each micrograph |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 4.0 Å / Resolution method: OTHER / Software - Name: RELION / Number images used: 246065 |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)



















 baculovirus-insect cells
baculovirus-insect cells