+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of Rpd3S in loose-state Rpd3S-NCP complex | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Dynamic Histone Modifications / Gene Regulation / Histone Deacetylase Complex | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationTINTIN complex / negative regulation of antisense RNA transcription / Snt2C complex / negative regulation of silent mating-type cassette heterochromatin formation / negative regulation of reciprocal meiotic recombination / Rpd3L complex / protein localization to nucleolar rDNA repeats / negative regulation of rDNA heterochromatin formation / Rpd3L-Expanded complex / Rpd3S complex ...TINTIN complex / negative regulation of antisense RNA transcription / Snt2C complex / negative regulation of silent mating-type cassette heterochromatin formation / negative regulation of reciprocal meiotic recombination / Rpd3L complex / protein localization to nucleolar rDNA repeats / negative regulation of rDNA heterochromatin formation / Rpd3L-Expanded complex / Rpd3S complex / rDNA chromatin condensation / nucleophagy / regulation of RNA stability / HDACs deacetylate histones / histone deacetylase activity, hydrolytic mechanism / DNA replication-dependent chromatin assembly / histone deacetylase / nucleosome disassembly / cellular response to nitrogen starvation / SUMOylation of chromatin organization proteins / regulation of DNA-templated DNA replication initiation / histone deacetylase activity / negative regulation of transcription by RNA polymerase I / histone deacetylase complex / : / Sin3-type complex / NuA4 histone acetyltransferase complex / histone reader activity / Estrogen-dependent gene expression / histone acetyltransferase complex / positive regulation of macroautophagy / nuclear periphery / meiotic cell cycle / transcription elongation by RNA polymerase II / positive regulation of transcription elongation by RNA polymerase II / G1/S transition of mitotic cell cycle / double-strand break repair via nonhomologous end joining / G2/M transition of mitotic cell cycle / transcription corepressor activity / heterochromatin formation / nucleosome assembly / cellular response to heat / response to oxidative stress / transcription coactivator activity / cell division / DNA repair / negative regulation of DNA-templated transcription / DNA-templated transcription / regulation of DNA-templated transcription / regulation of transcription by RNA polymerase II / chromatin / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / zinc ion binding / identical protein binding / nucleus / cytoplasm Similarity search - Function | |||||||||||||||
| Biological species | synthetic construct (others) /  | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||||||||
 Authors Authors | Li HT / Yan CY / Guan HP / Wang P | |||||||||||||||
| Funding support |  China, 4 items China, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S. Authors: Haipeng Guan / Pei Wang / Pei Zhang / Chun Ruan / Yutian Ou / Bo Peng / Xiangdong Zheng / Jianlin Lei / Bing Li / Chuangye Yan / Haitao Li /  Abstract: Context-dependent dynamic histone modifications constitute a key epigenetic mechanism in gene regulation. The Rpd3 small (Rpd3S) complex recognizes histone H3 trimethylation on lysine 36 (H3K36me3) ...Context-dependent dynamic histone modifications constitute a key epigenetic mechanism in gene regulation. The Rpd3 small (Rpd3S) complex recognizes histone H3 trimethylation on lysine 36 (H3K36me3) and deacetylates histones H3 and H4 at multiple sites across transcribed regions. Here we solved the cryo-electron microscopy structures of Saccharomyces cerevisiae Rpd3S in its free and H3K36me3 nucleosome-bound states. We demonstrated a unique architecture of Rpd3S, in which two copies of Eaf3-Rco1 heterodimers are asymmetrically assembled with Rpd3 and Sin3 to form a catalytic core complex. Multivalent recognition of two H3K36me3 marks, nucleosomal DNA and linker DNAs by Eaf3, Sin3 and Rco1 positions the catalytic centre of Rpd3 next to the histone H4 N-terminal tail for deacetylation. In an alternative catalytic mode, combinatorial readout of unmethylated histone H3 lysine 4 and H3K36me3 by Rco1 and Eaf3 directs histone H3-specific deacetylation except for the registered histone H3 acetylated lysine 9. Collectively, our work illustrates dynamic and diverse modes of multivalent nucleosomal engagement and methylation-guided deacetylation by Rpd3S, highlighting the exquisite complexity of epigenetic regulation with delicately designed multi-subunit enzymatic machineries in transcription and beyond. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33849.map.gz emd_33849.map.gz | 7.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33849-v30.xml emd-33849-v30.xml emd-33849.xml emd-33849.xml | 29.1 KB 29.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33849_fsc.xml emd_33849_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_33849.png emd_33849.png | 141.3 KB | ||
| Filedesc metadata |  emd-33849.cif.gz emd-33849.cif.gz | 8.8 KB | ||
| Others |  emd_33849_half_map_1.map.gz emd_33849_half_map_1.map.gz emd_33849_half_map_2.map.gz emd_33849_half_map_2.map.gz | 80.8 MB 80.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33849 http://ftp.pdbj.org/pub/emdb/structures/EMD-33849 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33849 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33849 | HTTPS FTP |
-Related structure data
| Related structure data |  7yi2MC  7yi0C  7yi1C  7yi3C  7yi4C  7yi5C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33849.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33849.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0825 Å | ||||||||||||||||||||||||||||||||||||
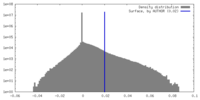
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_33849_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
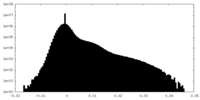
| Density Histograms |
-Half map: #2
| File | emd_33849_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
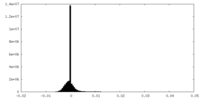
| Density Histograms |
- Sample components
Sample components
+Entire : Rpd3S complex with H3K36me3 nucleosome
+Supramolecule #1: Rpd3S complex with H3K36me3 nucleosome
+Supramolecule #2: DNA
+Supramolecule #3: SIN3/RPD3/EAF3/RCO1
+Macromolecule #1: Wisdom 601 DNA (167-MER)
+Macromolecule #2: Wisdom 601 DNA (167-MER)
+Macromolecule #3: Transcriptional regulatory protein SIN3
+Macromolecule #4: Histone deacetylase RPD3
+Macromolecule #5: Chromatin modification-related protein EAF3
+Macromolecule #6: Transcriptional regulatory protein RCO1
+Macromolecule #7: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 1.8 µm / Calibrated defocus min: 1.5 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)