+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of Rpd3S complex | |||||||||||||||
 Map data Map data | Cryo-EM map of Rpd3S complex | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Dynamic Histone Modifications / Gene Regulation / Histone Deacetylase Complex | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationTINTIN complex / negative regulation of antisense RNA transcription / Snt2C complex / negative regulation of silent mating-type cassette heterochromatin formation / negative regulation of reciprocal meiotic recombination / Rpd3L complex / protein localization to nucleolar rDNA repeats / negative regulation of rDNA heterochromatin formation / Rpd3L-Expanded complex / Rpd3S complex ...TINTIN complex / negative regulation of antisense RNA transcription / Snt2C complex / negative regulation of silent mating-type cassette heterochromatin formation / negative regulation of reciprocal meiotic recombination / Rpd3L complex / protein localization to nucleolar rDNA repeats / negative regulation of rDNA heterochromatin formation / Rpd3L-Expanded complex / Rpd3S complex / rDNA chromatin condensation / nucleophagy / regulation of RNA stability / HDACs deacetylate histones / histone deacetylase activity, hydrolytic mechanism / DNA replication-dependent chromatin assembly / histone deacetylase / nucleosome disassembly / cellular response to nitrogen starvation / SUMOylation of chromatin organization proteins / regulation of DNA-templated DNA replication initiation / histone deacetylase activity / negative regulation of transcription by RNA polymerase I / histone deacetylase complex / : / Sin3-type complex / NuA4 histone acetyltransferase complex / histone reader activity / Estrogen-dependent gene expression / histone acetyltransferase complex / positive regulation of macroautophagy / nuclear periphery / meiotic cell cycle / transcription elongation by RNA polymerase II / positive regulation of transcription elongation by RNA polymerase II / G1/S transition of mitotic cell cycle / double-strand break repair via nonhomologous end joining / G2/M transition of mitotic cell cycle / transcription corepressor activity / heterochromatin formation / nucleosome assembly / cellular response to heat / response to oxidative stress / transcription coactivator activity / cell division / DNA repair / negative regulation of DNA-templated transcription / DNA-templated transcription / regulation of DNA-templated transcription / regulation of transcription by RNA polymerase II / chromatin / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / zinc ion binding / identical protein binding / nucleus / cytoplasm Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||||||||
 Authors Authors | Li HT / Yan CY / Guan HP / Wang P | |||||||||||||||
| Funding support |  China, 4 items China, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S. Authors: Haipeng Guan / Pei Wang / Pei Zhang / Chun Ruan / Yutian Ou / Bo Peng / Xiangdong Zheng / Jianlin Lei / Bing Li / Chuangye Yan / Haitao Li /  Abstract: Context-dependent dynamic histone modifications constitute a key epigenetic mechanism in gene regulation. The Rpd3 small (Rpd3S) complex recognizes histone H3 trimethylation on lysine 36 (H3K36me3) ...Context-dependent dynamic histone modifications constitute a key epigenetic mechanism in gene regulation. The Rpd3 small (Rpd3S) complex recognizes histone H3 trimethylation on lysine 36 (H3K36me3) and deacetylates histones H3 and H4 at multiple sites across transcribed regions. Here we solved the cryo-electron microscopy structures of Saccharomyces cerevisiae Rpd3S in its free and H3K36me3 nucleosome-bound states. We demonstrated a unique architecture of Rpd3S, in which two copies of Eaf3-Rco1 heterodimers are asymmetrically assembled with Rpd3 and Sin3 to form a catalytic core complex. Multivalent recognition of two H3K36me3 marks, nucleosomal DNA and linker DNAs by Eaf3, Sin3 and Rco1 positions the catalytic centre of Rpd3 next to the histone H4 N-terminal tail for deacetylation. In an alternative catalytic mode, combinatorial readout of unmethylated histone H3 lysine 4 and H3K36me3 by Rco1 and Eaf3 directs histone H3-specific deacetylation except for the registered histone H3 acetylated lysine 9. Collectively, our work illustrates dynamic and diverse modes of multivalent nucleosomal engagement and methylation-guided deacetylation by Rpd3S, highlighting the exquisite complexity of epigenetic regulation with delicately designed multi-subunit enzymatic machineries in transcription and beyond. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33845.map.gz emd_33845.map.gz | 5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33845-v30.xml emd-33845-v30.xml emd-33845.xml emd-33845.xml | 26.3 KB 26.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33845_fsc.xml emd_33845_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_33845.png emd_33845.png | 134.5 KB | ||
| Filedesc metadata |  emd-33845.cif.gz emd-33845.cif.gz | 8.2 KB | ||
| Others |  emd_33845_half_map_1.map.gz emd_33845_half_map_1.map.gz emd_33845_half_map_2.map.gz emd_33845_half_map_2.map.gz | 49.4 MB 49.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33845 http://ftp.pdbj.org/pub/emdb/structures/EMD-33845 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33845 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33845 | HTTPS FTP |
-Related structure data
| Related structure data |  7yi0MC  7yi1C  7yi2C  7yi3C  7yi4C  7yi5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33845.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33845.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of Rpd3S complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
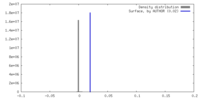
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_33845_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
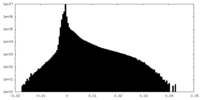
| Density Histograms |
-Half map: #2
| File | emd_33845_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
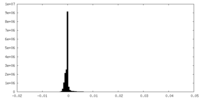
| Density Histograms |
- Sample components
Sample components
-Entire : Rpd3S complex
| Entire | Name: Rpd3S complex |
|---|---|
| Components |
|
-Supramolecule #1: Rpd3S complex
| Supramolecule | Name: Rpd3S complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 523 kDa/nm |
-Macromolecule #1: Transcriptional regulatory protein SIN3
| Macromolecule | Name: Transcriptional regulatory protein SIN3 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 175.047266 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSQVWHNSNS QSNDVATSND ATGSNERNEK EPSLQGNKPG FVQQQQRITL PSLSALSTKE EDRRDSNGQQ ALTSHAAHIL GYPPPHSNA MPSIATDSAL KQPHEYHPRP KSSSSSPSIN ASLMNAGPAP LPTVGAASFS LSRFDNPLPI KAPVHTEEPK S YNGLQEEE ...String: MSQVWHNSNS QSNDVATSND ATGSNERNEK EPSLQGNKPG FVQQQQRITL PSLSALSTKE EDRRDSNGQQ ALTSHAAHIL GYPPPHSNA MPSIATDSAL KQPHEYHPRP KSSSSSPSIN ASLMNAGPAP LPTVGAASFS LSRFDNPLPI KAPVHTEEPK S YNGLQEEE KATQRPQDCK EVPAGVQPAD APDPSSNHAD ANDDNNNNEN SHDEDADYRP LNVKDALSYL EQVKFQFSSR PD IYNLFLD IMKDFKSQAI DTPGVIERVS TLFRGYPILI QGFNTFLPQG YRIECSSNPD DPIRVTTPMG TTTVNNNISP SGR GTTDAQ ELGSFPESDG NGVQQPSNVP MVPSSVYQSE QNQDQQQSLP LLATSSGLPS IQQPEMPAHR QIPQSQSLVP QEDA KKNVD VEFSQAISYV NKIKTRFADQ PDIYKHFLEI LQTYQREQKP INEVYAQVTH LFQNAPDLLE DFKKFLPDSS ASANQ QVQH AQQHAQQQHE AQMHAQAQAQ AQAQAQVEQQ KQQQQFLYPA SGYYGHPSNR GIPQQNLPPI GSFSPPTNGS TVHEAY QDQ QHMQPPHFMP LPSIVQHGPN MVHQGIANEN PPLSDLRTSL TEQYAPSSIQ HQQQHPQSIS PIANTQYGDI PVRPEID LD PSIVPVVPEP TEPIENNISL NEEVTFFEKA KRYIGNKHLY TEFLKILNLY SQDILDLDDL VEKVDFYLGS NKELFTWF K NFVGYQEKTK CIENIVHEKH RLDLDLCEAF GPSYKRLPKS DTFMPCSGRD DMCWEVLNDE WVGHPVWASE DSGFIAHRK NQYEETLFKI EEERHEYDFY IESNLRTIQC LETIVNKIEN MTENEKANFK LPPGLGHTSM TIYKKVIRKV YDKERGFEII DALHEHPAV TAPVVLKRLK QKDEEWRRAQ REWNKVWREL EQKVFFKSLD HLGLTFKQAD KKLLTTKQLI SEISSIKVDQ T NKKIHWLT PKPKSQLDFD FPDKNIFYDI LCLADTFITH TTAYSNPDKE RLKDLLKYFI SLFFSISFEK IEESLYSHKQ NV SESSGSD DGSSIASRKR PYQQEMSLLD ILHRSRYQKL KRSNDEDGKV PQLSEPPEEE PNTIEEEELI DEEAKNPWLT GNL VEEANS QGIIQNRSIF NLFANTNIYI FFRHWTTIYE RLLEIKQMNE RVTKEINTRS TVTFAKDLDL LSSQLSEMGL DFVG EDAYK QVLRLSRRLI NGDLEHQWFE ESLRQAYNNK AFKLYTIDKV TQSLVKHAHT LMTDAKTAEI MALFVKDRNA STTSA KDQI IYRLQVRSHM SNTENMFRIE FDKRTLHVSI QYIALDDLTL KEPKADEDKW KYYVTSYALP HPTEGIPHEK LKIPFL ERL IEFGQDIDGT EVDEEFSPEG ISVSTLKIKI QPITYQLHIE NGSYDVFTRK ATNKYPTIAN DNTQKGMVSQ KKELISK FL DCAVGLRNNL DEAQKLSMQK KWENLKDSIA KTSAGNQGIE SETEKGKITK QEQSDNLDSS TASVLPASIT TVPQDDNI E TTGNTESSDK GAKIQ UniProtKB: Transcriptional regulatory protein SIN3 |
-Macromolecule #2: Histone deacetylase RPD3
| Macromolecule | Name: Histone deacetylase RPD3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: histone deacetylase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 48.961957 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVYEATPFDP ITVKPSDKRR VAYFYDADVG NYAYGAGHPM KPHRIRMAHS LIMNYGLYKK MEIYRAKPAT KQEMCQFHTD EYIDFLSRV TPDNLEMFKR ESVKFNVGDD CPVFDGLYEY CSISGGGSME GAARLNRGKC DVAVNYAGGL HHAKKSEASG F CYLNDIVL ...String: MVYEATPFDP ITVKPSDKRR VAYFYDADVG NYAYGAGHPM KPHRIRMAHS LIMNYGLYKK MEIYRAKPAT KQEMCQFHTD EYIDFLSRV TPDNLEMFKR ESVKFNVGDD CPVFDGLYEY CSISGGGSME GAARLNRGKC DVAVNYAGGL HHAKKSEASG F CYLNDIVL GIIELLRYHP RVLYIDIDVH HGDGVEEAFY TTDRVMTCSF HKYGEFFPGT GELRDIGVGA GKNYAVNVPL RD GIDDATY RSVFEPVIKK IMEWYQPSAV VLQCGGDSLS GDRLGCFNLS MEGHANCVNY VKSFGIPMMV VGGGGYTMRN VAR TWCFET GLLNNVVLDK DLPYNEYYEY YGPDYKLSVR PSNMFNVNTP EYLDKVMTNI FANLENTKYA PSVQLNHTPR DAED LGDVE EDSAEAKDTK GGSQYARDLH VEHDNEFY UniProtKB: Histone deacetylase RPD3 |
-Macromolecule #3: Chromatin modification-related protein EAF3
| Macromolecule | Name: Chromatin modification-related protein EAF3 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.266406 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVDLEQEFAL GGRCLAFHGP LMYEAKILKI WDPSSKMYTS IPNDKPGGSS QATKEIKPQK LGEDESIPEE IINGKCFFIH YQGWKSSWD EWVGYDRIRA YNEENIAMKK RLANEAKEAK KSLLEQQKKK KLSTSLGGPS NGGKRKGDSR SNASISKSTS Q SFLTSSVS ...String: MVDLEQEFAL GGRCLAFHGP LMYEAKILKI WDPSSKMYTS IPNDKPGGSS QATKEIKPQK LGEDESIPEE IINGKCFFIH YQGWKSSWD EWVGYDRIRA YNEENIAMKK RLANEAKEAK KSLLEQQKKK KLSTSLGGPS NGGKRKGDSR SNASISKSTS Q SFLTSSVS GRKSGRSSAN SLHPGSSLRS SSDQNGNDDR RRSSSLSPNM LHHIAGYPTP KISLQIPIKL KSVLVDDWEY VT KDKKICR LPADVTVEMV LNKYEHEVSQ ELESPGSQSQ LSEYCAGLKL YFDKCLGNML LYRLERLQYD ELLKKSSKDQ KPL VPIRIY GAIHLLRLIS VLPELISSTT MDLQSCQLLI KQTEDFLVWL LMHVDEYFND KDPNRSDDAL YVNTSSQYEG VALG M UniProtKB: Chromatin modification-related protein EAF3 |
-Macromolecule #4: Transcriptional regulatory protein RCO1
| Macromolecule | Name: Transcriptional regulatory protein RCO1 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 78.951305 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDTSKKDTTR SPSHSNSSSP SSSSLSSSSS KEKKRPKRLS SQNVNYDLKR RKIITSEGIE RSFKNEHSNL AVEDNIPEEE PKELLEKDS KGNIIKLNEP STISEDSKVS VTGLPLNKGP SEKIKRESLW NYRKNLGGQS NNSEMTLVPS KRFTQVPKNF Q DLNRNDLK ...String: MDTSKKDTTR SPSHSNSSSP SSSSLSSSSS KEKKRPKRLS SQNVNYDLKR RKIITSEGIE RSFKNEHSNL AVEDNIPEEE PKELLEKDS KGNIIKLNEP STISEDSKVS VTGLPLNKGP SEKIKRESLW NYRKNLGGQS NNSEMTLVPS KRFTQVPKNF Q DLNRNDLK TFLTENMTEE SNIRSTIGWN GDIINRTRDR EPESDRDNKK LSNIRTKIIL STNATYDSKS KLFGQNSIKS TS NASEKIF RDKNNSTIDF ENEDFCSACN QSGSFLCCDT CPKSFHFLCL DPPIDPNNLP KGDWHCNECK FKIFINNSMA TLK KIESNF IKQNNNVKIF AKLLFNIDSH NPKQFQLPNY IKETFPAVKT GSRGQYSDEN DKIPLTDRQL FNTSYGQSIT KLDS YNPDT HIDSNSGKFL ICYKCNQTRL GSWSHPENSR LIMTCDYCQT PWHLDCVPRA SFKNLGSKWK CPLHSPTKVY KKIHH CQED NSVNYKVWKK QRLINKKNQL YYEPLQKIGY QNNGNIQIIP TTSHTDYDFN QDFKITQIDE NSIKYDFFDK IYKSKM VQK RKLFQFQESL IDKLVSNGSQ NGNSEDNMVK DIASLIYFQV SNNDKSSNNK SASKSNNLRK LWDLKELTNV VVPNELD SI QFNDFSSDEI KHLLYLKKII ESKPKEELLK FLNIENPENQ SE UniProtKB: Transcriptional regulatory protein RCO1 |
-Macromolecule #5: Transcriptional regulatory protein RCO1
| Macromolecule | Name: Transcriptional regulatory protein RCO1 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 78.875211 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDTSKKDTTR SPSHSNSSSP SSSSLSSSSS KEKKRPKRLS SQNVNYDLKR RKIITSEGIE RSFKNEHSNL AVEDNIPEEE PKELLEKDS KGNIIKLNEP STISEDSKVS VTGLPLNKGP SEKIKRESLW NYRKNLGGQS NNSEMTLVPS KRFTQVPKNF Q DLNRNDLK ...String: MDTSKKDTTR SPSHSNSSSP SSSSLSSSSS KEKKRPKRLS SQNVNYDLKR RKIITSEGIE RSFKNEHSNL AVEDNIPEEE PKELLEKDS KGNIIKLNEP STISEDSKVS VTGLPLNKGP SEKIKRESLW NYRKNLGGQS NNSEMTLVPS KRFTQVPKNF Q DLNRNDLK TFLTENMTEE SNIRSTIGWN GDIINRTRDR EPESDRDNKK LSNIRTKIIL STNATYDSKS KLFGQNSIKS TS NASEKIF RDKNNSTIDF ENEDFCSACN QSGSFLCCDT CPKSFHFLCL DPPIDPNNLP KGDWHCNECK FKIFINNSMA TLK KIESNF IKQNNNVKIF AKLLFNIDSH NPKQFQLPNY IKETFPAVKT GSRGQYSDEN DKIPLTDRQL FNTSYGQSIT KLDS YNPDT HIDSNSGKFL ICYKCNQTRL GSWSHPENSR LIMTCDYCQT PWHLDCVPRA SFKNLGSKWK CPLHSPTKVY KKIHH CQED NSVNYKVWKK QRLINKKNQL YYEPLQKIGY QNNGNIQIIP TTSHTDYDFN QDFKITQIDE NSIKYDFFDK IYKSKM VQK RKLAQFQESL IDKLVSNGSQ NGNSEDNMVK DIASLIYFQV SNNDKSSNNK SASKSNNLRK LWDLKELTNV VVPNELD SI QFNDFSSDEI KHLLYLKKII ESKPKEELLK FLNIENPENQ SE UniProtKB: Transcriptional regulatory protein RCO1 |
-Macromolecule #6: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 6 / Number of copies: 7 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 1.8 µm / Calibrated defocus min: 1.5 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)