[English] 日本語
 Yorodumi
Yorodumi- EMDB-3362: Natively membrane-anchored full-length Herpes simplex virus 1 gly... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3362 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Natively membrane-anchored full-length Herpes simplex virus 1 glycoprotein B | |||||||||
 Map data Map data | Subtomogram average of HSV-1 gB on membrane | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | membrane fusion / class III fusogen / pre-fusion | |||||||||
| Function / homology |  Function and homology information Function and homology informationvirion component => GO:0044423 / host cell Golgi membrane / viral process / host cell membrane / membrane => GO:0016020 / host cell endosome membrane / host cell endosome / host cell Golgi apparatus / viral envelope / symbiont entry into host cell ...virion component => GO:0044423 / host cell Golgi membrane / viral process / host cell membrane / membrane => GO:0016020 / host cell endosome membrane / host cell endosome / host cell Golgi apparatus / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / virion membrane / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |   Human herpesvirus 1 (Herpes simplex virus type 1) Human herpesvirus 1 (Herpes simplex virus type 1) | |||||||||
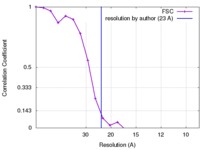
| Method | subtomogram averaging / cryo EM / Resolution: 23.0 Å | |||||||||
 Authors Authors | Zeev-Ben-Mordehai T / Vasishtan D / Duran AH / Vollmer B / White P / Pandurangan AP / Siebert CA / Topf M / Grunewald K | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2016 Journal: Proc Natl Acad Sci U S A / Year: 2016Title: Two distinct trimeric conformations of natively membrane-anchored full-length herpes simplex virus 1 glycoprotein B. Authors: Tzviya Zeev-Ben-Mordehai / Daven Vasishtan / Anna Hernández Durán / Benjamin Vollmer / Paul White / Arun Prasad Pandurangan / C Alistair Siebert / Maya Topf / Kay Grünewald /  Abstract: Many viruses are enveloped by a lipid bilayer acquired during assembly, which is typically studded with one or two types of glycoproteins. These viral surface proteins act as the primary interface ...Many viruses are enveloped by a lipid bilayer acquired during assembly, which is typically studded with one or two types of glycoproteins. These viral surface proteins act as the primary interface between the virus and the host. Entry of enveloped viruses relies on specialized fusogen proteins to help merge the virus membrane with the host membrane. In the multicomponent herpesvirus fusion machinery, glycoprotein B (gB) acts as this fusogen. Although the structure of the gB ectodomain postfusion conformation has been determined, any other conformations (e.g., prefusion, intermediate conformations) have so far remained elusive, thus restricting efforts to develop antiviral treatments and prophylactic vaccines. Here, we have characterized the full-length herpes simplex virus 1 gB in a native membrane by displaying it on cell-derived vesicles and using electron cryotomography. Alongside the known postfusion conformation, a novel one was identified. Its structure, in the context of the membrane, was determined by subvolume averaging and found to be trimeric like the postfusion conformation, but appeared more condensed. Hierarchical constrained density-fitting of domains unexpectedly revealed the fusion loops in this conformation to be apart and pointing away from the anchoring membrane. This vital observation is a substantial step forward in understanding the complex herpesvirus fusion mechanism, and opens up new opportunities for more targeted intervention of herpesvirus entry. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3362.map.gz emd_3362.map.gz | 1.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3362-v30.xml emd-3362-v30.xml emd-3362.xml emd-3362.xml | 11.9 KB 11.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3362_fsc.xml emd_3362_fsc.xml | 2 KB | Display |  FSC data file FSC data file |
| Images |  3362_emdb_thumb.png 3362_emdb_thumb.png | 102.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3362 http://ftp.pdbj.org/pub/emdb/structures/EMD-3362 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3362 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3362 | HTTPS FTP |
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3362.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3362.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subtomogram average of HSV-1 gB on membrane | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.6 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Glycoprotein B of Herpes Simplex Virus 1
| Entire | Name: Glycoprotein B of Herpes Simplex Virus 1 |
|---|---|
| Components |
|
-Supramolecule #1000: Glycoprotein B of Herpes Simplex Virus 1
| Supramolecule | Name: Glycoprotein B of Herpes Simplex Virus 1 / type: sample / ID: 1000 / Details: Sample bound to extracellular vesicles / Oligomeric state: Trimeric / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 300 KDa |
-Macromolecule #1: Glycoprotein B
| Macromolecule | Name: Glycoprotein B / type: protein_or_peptide / ID: 1 / Name.synonym: gB / Number of copies: 3 / Oligomeric state: Trimeric / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Human herpesvirus 1 (Herpes simplex virus type 1) / synonym: HSV-1 Human herpesvirus 1 (Herpes simplex virus type 1) / synonym: HSV-1 |
| Molecular weight | Theoretical: 300 KDa |
| Recombinant expression | Organism:  Mesocricetus auratus (golden hamster) / Recombinant cell: BHK 21 / Recombinant plasmid: pPEP98 Mesocricetus auratus (golden hamster) / Recombinant cell: BHK 21 / Recombinant plasmid: pPEP98 |
| Sequence | UniProtKB: Envelope glycoprotein B GO: viral process, virion attachment to host cell, symbiont entry into host cell, membrane, membrane => GO:0016020, virion component => GO:0044423, viral envelope, host cell plasma membrane, host ...GO: viral process, virion attachment to host cell, symbiont entry into host cell, membrane, membrane => GO:0016020, virion component => GO:0044423, viral envelope, host cell plasma membrane, host cell membrane, host cell endosome, host cell endosome membrane, host cell Golgi apparatus, host cell Golgi membrane, virion membrane InterPro: Herpesvirus Glycoprotein B |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: 25mM HEPES, 130mM NaCl |
|---|---|
| Grid | Details: Holey carbon on top of 200 mesh gold grid. |
| Vitrification | Cryogen name: ETHANE-PROPANE MIXTURE / Chamber temperature: 77 K / Instrument: HOMEMADE PLUNGER / Method: Blot for 3 sec before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Min: 80 K / Max: 100 K / Average: 85 K |
| Specialist optics | Energy filter - Name: Gatan Quantum 964 / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 20.0 eV |
| Details | counting mode |
| Date | Sep 16, 2014 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Number real images: 20 / Average electron dose: 60 e/Å2 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 6.0 µm / Nominal magnification: 95000 |
| Sample stage | Specimen holder: Polara holder / Specimen holder model: GATAN LIQUID NITROGEN / Tilt series - Axis1 - Min angle: -52 ° / Tilt series - Axis1 - Max angle: 53 ° |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Software | Name: Chimera, Flex-EM, ADP-EM, GMFit, TEMPy |
| Details | Individual domains of 2GUM were fitted separately, using a hierarchical fitting approach |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: CCC, SCCC, Atom Protrusion Score |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)