[English] 日本語
 Yorodumi
Yorodumi- EMDB-33447: RNA polymerase II elongation complex transcribing a nucleosome (E... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | RNA polymerase II elongation complex transcribing a nucleosome (EC58oct) | ||||||||||||||||||||||||
 Map data Map data | |||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | chromatin / nucleosome / TRANSCRIPTION / TRANSCRIPTION-DNA-RNA complex | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationconstitutive heterochromatin formation / FACT complex / regulation of septum digestion after cytokinesis / RNA polymerase II C-terminal domain phosphoserine binding / co-transcriptional lncRNA 3' end processing, cleavage and polyadenylation pathway / Cdc73/Paf1 complex / siRNA-mediated pericentric heterochromatin formation / histone chaperone activity / negative regulation of chromosome condensation / Barr body ...constitutive heterochromatin formation / FACT complex / regulation of septum digestion after cytokinesis / RNA polymerase II C-terminal domain phosphoserine binding / co-transcriptional lncRNA 3' end processing, cleavage and polyadenylation pathway / Cdc73/Paf1 complex / siRNA-mediated pericentric heterochromatin formation / histone chaperone activity / negative regulation of chromosome condensation / Barr body / : / DSIF complex / RPB4-RPB7 complex / pericentric heterochromatin formation / inner kinetochore / nucleosome organization / muscle cell differentiation / transcription elongation factor activity / intracellular phosphate ion homeostasis / chromatin-protein adaptor activity / oocyte maturation / nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay / nucleosomal DNA binding / transcription elongation-coupled chromatin remodeling / termination of RNA polymerase II transcription / poly(A)+ mRNA export from nucleus / termination of RNA polymerase III transcription / positive regulation of nuclear-transcribed mRNA poly(A) tail shortening / nucleus organization / transcription initiation at RNA polymerase III promoter / termination of RNA polymerase I transcription / transcription initiation at RNA polymerase I promoter / RNA polymerase II complex binding / maintenance of transcriptional fidelity during transcription elongation by RNA polymerase II / positive regulation of translational initiation / nuclear-transcribed mRNA catabolic process / spermatid development / chromosome, centromeric region / negative regulation of tumor necrosis factor-mediated signaling pathway / single fertilization / subtelomeric heterochromatin formation / translation elongation factor activity / RNA polymerase I complex / RNA polymerase III complex / transcription elongation by RNA polymerase I / RNA polymerase II core promoter sequence-specific DNA binding / pericentric heterochromatin / translesion synthesis / RNA polymerase II, core complex / tRNA transcription by RNA polymerase III / nucleosome binding / transcription by RNA polymerase I / negative regulation of megakaryocyte differentiation / protein localization to CENP-A containing chromatin / Replacement of protamines by nucleosomes in the male pronucleus / CENP-A containing nucleosome / transcription-coupled nucleotide-excision repair / translation initiation factor binding / Packaging Of Telomere Ends / Recognition and association of DNA glycosylase with site containing an affected purine / Cleavage of the damaged purine / embryo implantation / Deposition of new CENPA-containing nucleosomes at the centromere / telomere organization / Recognition and association of DNA glycosylase with site containing an affected pyrimidine / Cleavage of the damaged pyrimidine / RNA Polymerase I Promoter Opening / Inhibition of DNA recombination at telomere / Assembly of the ORC complex at the origin of replication / Meiotic synapsis / SUMOylation of chromatin organization proteins / Regulation of endogenous retroelements by the Human Silencing Hub (HUSH) complex / regulation of DNA-templated transcription elongation / DNA methylation / transcription elongation factor complex / Condensation of Prophase Chromosomes / Chromatin modifications during the maternal to zygotic transition (MZT) / SIRT1 negatively regulates rRNA expression / HCMV Late Events / ERCC6 (CSB) and EHMT2 (G9a) positively regulate rRNA expression / PRC2 methylates histones and DNA / Regulation of endogenous retroelements by KRAB-ZFP proteins / innate immune response in mucosa / Defective pyroptosis / Negative Regulation of CDH1 Gene Transcription / transcription initiation at RNA polymerase II promoter / HDACs deacetylate histones / transcription elongation by RNA polymerase II / Regulation of endogenous retroelements by Piwi-interacting RNAs (piRNAs) / P-body / positive regulation of transcription elongation by RNA polymerase II / Nonhomologous End-Joining (NHEJ) / RNA Polymerase I Promoter Escape / lipopolysaccharide binding / Transcriptional regulation by small RNAs / Formation of the beta-catenin:TCF transactivating complex / Activated PKN1 stimulates transcription of AR (androgen receptor) regulated genes KLK2 and KLK3 / HDMs demethylate histones / RUNX1 regulates genes involved in megakaryocyte differentiation and platelet function / G2/M DNA damage checkpoint Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  Komagataella phaffii (fungus) / Komagataella phaffii (fungus) /  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | ||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | ||||||||||||||||||||||||
 Authors Authors | Ehara H / Kujirai T / Shirouzu M / Kurumizaka H / Sekine S | ||||||||||||||||||||||||
| Funding support |  Japan, 7 items Japan, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT. Authors: Haruhiko Ehara / Tomoya Kujirai / Mikako Shirouzu / Hitoshi Kurumizaka / Shun-Ichi Sekine /  Abstract: During gene transcription, RNA polymerase II (RNAPII) traverses nucleosomes in chromatin, but the mechanism has remained elusive. Using cryo-electron microscopy, we obtained structures of the RNAPII ...During gene transcription, RNA polymerase II (RNAPII) traverses nucleosomes in chromatin, but the mechanism has remained elusive. Using cryo-electron microscopy, we obtained structures of the RNAPII elongation complex (EC) passing through a nucleosome in the presence of the transcription elongation factors Spt6, Spn1, Elf1, Spt4/5, and Paf1C and the histone chaperone FACT (facilitates chromatin transcription). The structures show snapshots of EC progression on DNA mediating downstream nucleosome disassembly, followed by its reassembly upstream of the EC, which is facilitated by FACT. FACT dynamically adapts to successively occurring subnucleosome intermediates, forming an interface with the EC. Spt6, Spt4/5, and Paf1C form a "cradle" at the EC DNA-exit site and support the upstream nucleosome reassembly. These structures explain the mechanism by which the EC traverses nucleosomes while maintaining the chromatin structure and epigenetic information. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33447.map.gz emd_33447.map.gz | 40.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33447-v30.xml emd-33447-v30.xml emd-33447.xml emd-33447.xml | 67.7 KB 67.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33447_fsc.xml emd_33447_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_33447.png emd_33447.png | 102.8 KB | ||
| Filedesc metadata |  emd-33447.cif.gz emd-33447.cif.gz | 16.4 KB | ||
| Others |  emd_33447_additional_1.map.gz emd_33447_additional_1.map.gz emd_33447_additional_2.map.gz emd_33447_additional_2.map.gz emd_33447_half_map_1.map.gz emd_33447_half_map_1.map.gz emd_33447_half_map_2.map.gz emd_33447_half_map_2.map.gz | 40.6 MB 40.8 MB 40.9 MB 40.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33447 http://ftp.pdbj.org/pub/emdb/structures/EMD-33447 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33447 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33447 | HTTPS FTP |
-Related structure data
| Related structure data |  7xtdMC  7xn7C  7xseC  7xsxC  7xszC  7xt7C  7xtiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33447.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33447.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.484 Å | ||||||||||||||||||||||||||||||||||||
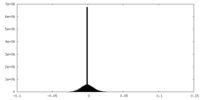
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: nucleosome reconstruction
| File | emd_33447_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | nucleosome reconstruction | ||||||||||||
| Projections & Slices |
| ||||||||||||
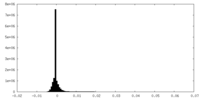
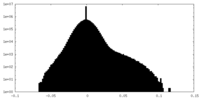
| Density Histograms |
-Additional map: EC reconstruction
| File | emd_33447_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EC reconstruction | ||||||||||||
| Projections & Slices |
| ||||||||||||
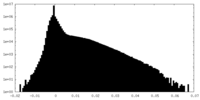
| Density Histograms |
-Half map: #1
| File | emd_33447_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
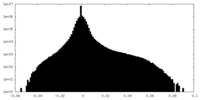
| Density Histograms |
-Half map: #2
| File | emd_33447_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
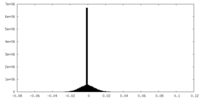
| Density Histograms |
- Sample components
Sample components
+Entire : RNA polymerase II elongation complex transcribing a nucleosome (E...
+Supramolecule #1: RNA polymerase II elongation complex transcribing a nucleosome (E...
+Supramolecule #2: RNA polymerase II
+Supramolecule #3: Elf1,Spt4/5,Spn1 and Paf1C
+Supramolecule #4: DNA,RNA
+Supramolecule #5: histone, FACT complex subunit
+Supramolecule #6: Spt6
+Macromolecule #1: DNA-directed RNA polymerase subunit
+Macromolecule #2: DNA-directed RNA polymerase subunit beta
+Macromolecule #3: RNA polymerase II third largest subunit B44, part of central core
+Macromolecule #4: RNA polymerase II subunit B32
+Macromolecule #5: DNA-directed RNA polymerases I, II, and III subunit RPABC1
+Macromolecule #6: RNA polymerase subunit ABC23, common to RNA polymerases I, II, and III
+Macromolecule #7: RNA polymerase II subunit
+Macromolecule #8: DNA-directed RNA polymerases I, II, and III subunit RPABC3
+Macromolecule #9: DNA-directed RNA polymerase subunit
+Macromolecule #10: RNA polymerase subunit ABC10-beta, common to RNA polymerases I, I...
+Macromolecule #11: RNA polymerase II subunit B12.5
+Macromolecule #12: RNA polymerase subunit ABC10-alpha
+Macromolecule #13: Transcription elongation factor 1 homolog
+Macromolecule #17: Transcription elongation factor SPT4
+Macromolecule #18: Transcription elongation factor SPT5
+Macromolecule #19: Transcription elongation factor Spt6
+Macromolecule #20: Protein that interacts with Spt6p and copurifies with Spt5p and R...
+Macromolecule #21: Component of the Paf1p complex
+Macromolecule #22: RNAPII-associated chromatin remodeling Paf1 complex subunit
+Macromolecule #23: Leo1
+Macromolecule #24: RNAP II-associated protein
+Macromolecule #25: Constituent of Paf1 complex with RNA polymerase II, Paf1p, Hpr1p,...
+Macromolecule #26: Histone H3.3
+Macromolecule #27: Histone H4
+Macromolecule #28: Histone H2A type 1-B/E
+Macromolecule #29: Histone H2B type 1-J
+Macromolecule #30: FACT complex subunit
+Macromolecule #31: FACT complex subunit POB3
+Macromolecule #14: DNA (198-MER)
+Macromolecule #16: DNA (198-MER)
+Macromolecule #15: RNA (5'-R(P*UP*GP*UP*AP*AP*UP*CP*CP*CP*CP*UP*UP*GP*GP*CP*GP*GP*UP...
+Macromolecule #32: ZINC ION
+Macromolecule #33: MAGNESIUM ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 51.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)