[English] 日本語
 Yorodumi
Yorodumi- EMDB-33399: Consensus map of connexin43/Cx43/GJA1 gap junction intercellular ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Consensus map of connexin43/Cx43/GJA1 gap junction intercellular channel in LMNG/CHS detergents at pH ~6.9 | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
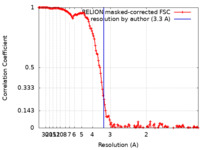
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Lee HJ / Cha HJ / Jeong H / Lee SN / Lee CW / Woo JS | |||||||||
| Funding support |  Korea, Republic Of, 1 items Korea, Republic Of, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM. Authors: Hyuk-Joon Lee / Hyung Jin Cha / Hyeongseop Jeong / Seu-Na Lee / Chang-Won Lee / Minsoo Kim / Jejoong Yoo / Jae-Sung Woo /  Abstract: Connexin family proteins assemble into hexameric hemichannels in the cell membrane. The hemichannels dock together between two adjacent membranes to form gap junction intercellular channels (GJIChs). ...Connexin family proteins assemble into hexameric hemichannels in the cell membrane. The hemichannels dock together between two adjacent membranes to form gap junction intercellular channels (GJIChs). We report the cryo-electron microscopy structures of Cx43 GJICh, revealing the dynamic equilibrium state of various channel conformations in detergents and lipid nanodiscs. We identify three different N-terminal helix conformations of Cx43-gate-covering (GCN), pore-lining (PLN), and flexible intermediate (FIN)-that are randomly distributed in purified GJICh particles. The conformational equilibrium shifts to GCN by cholesteryl hemisuccinates and to PLN by C-terminal truncations and at varying pH. While GJIChs that mainly comprise GCN protomers are occluded by lipids, those containing conformationally heterogeneous protomers show markedly different pore sizes. We observe an α-to-π-helix transition in the first transmembrane helix, which creates a side opening to the membrane in the FIN and PLN conformations. This study provides basic structural information to understand the mechanisms of action and regulation of Cx43 GJICh. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33399.map.gz emd_33399.map.gz | 27.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33399-v30.xml emd-33399-v30.xml emd-33399.xml emd-33399.xml | 13.4 KB 13.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33399_fsc.xml emd_33399_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_33399.png emd_33399.png | 38.5 KB | ||
| Others |  emd_33399_half_map_1.map.gz emd_33399_half_map_1.map.gz emd_33399_half_map_2.map.gz emd_33399_half_map_2.map.gz | 192.4 MB 192.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33399 http://ftp.pdbj.org/pub/emdb/structures/EMD-33399 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33399 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33399 | HTTPS FTP |
-Validation report
| Summary document |  emd_33399_validation.pdf.gz emd_33399_validation.pdf.gz | 892.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33399_full_validation.pdf.gz emd_33399_full_validation.pdf.gz | 892.4 KB | Display | |
| Data in XML |  emd_33399_validation.xml.gz emd_33399_validation.xml.gz | 21.1 KB | Display | |
| Data in CIF |  emd_33399_validation.cif.gz emd_33399_validation.cif.gz | 27.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33399 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33399 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33399 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33399 | HTTPS FTP |
-Related structure data
| Related structure data |  7f92C  7f93C  7f94C  7xq9C  7xqbC  7xqdC  7xqfC  7xqgC  7xqhC  7xqiC  7xqjC C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33399.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33399.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.675 Å | ||||||||||||||||||||||||||||||||||||
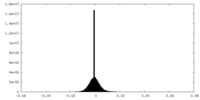
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map
| File | emd_33399_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
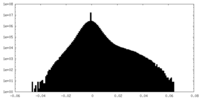
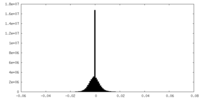
| Density Histograms |
-Half map: half map
| File | emd_33399_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
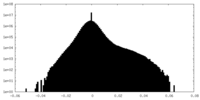
| Density Histograms |
- Sample components
Sample components
-Entire : Dodecameric complex of human Cx43/GJA1 in detergents (LMNG/CHS) a...
| Entire | Name: Dodecameric complex of human Cx43/GJA1 in detergents (LMNG/CHS) at pH ~6.9 |
|---|---|
| Components |
|
-Supramolecule #1: Dodecameric complex of human Cx43/GJA1 in detergents (LMNG/CHS) a...
| Supramolecule | Name: Dodecameric complex of human Cx43/GJA1 in detergents (LMNG/CHS) at pH ~6.9 type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all Details: Human Cx43 gap junction channel composed of two different protomers in pore-lining and gate-covering NTH conformations |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Gap junction alpha-1 protein (Cx43)
| Macromolecule | Name: Gap junction alpha-1 protein (Cx43) / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGDWSALGKL LDKVQAYSTA GGKVWLSVLF IFRILLLGTA VESAWGDEQS AFRCNTQQPG CENVCYDKSF PISHVRFWVL QIIFVSVPTL LYLAHVFYVM RKEEKLNKKE EELKVAQTDG VNVDMHLKQI EIKKFKYGIE EHGKVKMRGG LLRTYIISIL FKSIFEVAFL ...String: MGDWSALGKL LDKVQAYSTA GGKVWLSVLF IFRILLLGTA VESAWGDEQS AFRCNTQQPG CENVCYDKSF PISHVRFWVL QIIFVSVPTL LYLAHVFYVM RKEEKLNKKE EELKVAQTDG VNVDMHLKQI EIKKFKYGIE EHGKVKMRGG LLRTYIISIL FKSIFEVAFL LIQWYIYGFS LSAVYTCKRD PCPHQVDCFL SRPTEKTIFI IFMLVVSLVS LALNIIELFY VFFKGVKDRV KGKSDPYHAT SGALSPAKDC GSQKYAYFNG CSSPTAPLSP MSPPGYKLVT GDRNNSSCRN YNKQASEQNW ANYSAEQNRM GQAGSTISNS HAQPFDFPDD NQNSKKLAAG HELQPLAIVD QRPSSRASSR ASSRPRPDDL EI |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.9 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.75 µm / Nominal defocus min: 1.25 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)