[English] 日本語
 Yorodumi
Yorodumi- EMDB-33265: F1 domain of FoF1-ATPase with the up form of epsilon subunit from... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | F1 domain of FoF1-ATPase with the up form of epsilon subunit from Bacillus PS3 | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | ATP synthase F1 ATPase FoF1 / MOTOR PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationproton motive force-driven plasma membrane ATP synthesis / H+-transporting two-sector ATPase / proton-transporting ATP synthase complex / proton-transporting ATP synthase activity, rotational mechanism / ADP binding / ATP hydrolysis activity / ATP binding / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
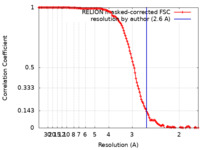
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||||||||
 Authors Authors | Nakano A / Kishikawa J / Nakanishi A / Mitsuoka K / Yokoyama K | |||||||||||||||
| Funding support |  Japan, 4 items Japan, 4 items
| |||||||||||||||
 Citation Citation |  Journal: PNAS Nexus / Year: 2022 Journal: PNAS Nexus / Year: 2022Title: Structural basis of unisite catalysis of bacterial FF-ATPase. Authors: Atsuki Nakano / Jun-Ichi Kishikawa / Atsuko Nakanishi / Kaoru Mitsuoka / Ken Yokoyama /  Abstract: Adenosine triphosphate (ATP) synthases (FF-ATPases) are crucial for all aerobic organisms. F, a water-soluble domain, can catalyze both the synthesis and hydrolysis of ATP with the rotation of the ...Adenosine triphosphate (ATP) synthases (FF-ATPases) are crucial for all aerobic organisms. F, a water-soluble domain, can catalyze both the synthesis and hydrolysis of ATP with the rotation of the central rotor inside a cylinder made of in three different conformations (referred to as , , and ). In this study, we determined multiple cryo-electron microscopy structures of bacterial FF exposed to different reaction conditions. The structures of nucleotide-depleted FF indicate that the ε subunit directly forces to adopt a closed form independent of the nucleotide binding to . The structure of FF under conditions that permit only a single catalytic subunit per enzyme to bind ATP is referred to as unisite catalysis and reveals that ATP hydrolysis unexpectedly occurs on instead of , where ATP hydrolysis proceeds in the steady-state catalysis of FF. This indicates that the unisite catalysis of bacterial FF significantly differs from the kinetics of steady-state turnover with continuous rotation of the shaft. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33265.map.gz emd_33265.map.gz | 140.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33265-v30.xml emd-33265-v30.xml emd-33265.xml emd-33265.xml | 23.5 KB 23.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33265_fsc.xml emd_33265_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_33265.png emd_33265.png | 78.4 KB | ||
| Masks |  emd_33265_msk_1.map emd_33265_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-33265.cif.gz emd-33265.cif.gz | 6.8 KB | ||
| Others |  emd_33265_additional_1.map.gz emd_33265_additional_1.map.gz emd_33265_half_map_1.map.gz emd_33265_half_map_1.map.gz emd_33265_half_map_2.map.gz emd_33265_half_map_2.map.gz | 166.5 MB 140.8 MB 140.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33265 http://ftp.pdbj.org/pub/emdb/structures/EMD-33265 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33265 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33265 | HTTPS FTP |
-Validation report
| Summary document |  emd_33265_validation.pdf.gz emd_33265_validation.pdf.gz | 890.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33265_full_validation.pdf.gz emd_33265_full_validation.pdf.gz | 890.4 KB | Display | |
| Data in XML |  emd_33265_validation.xml.gz emd_33265_validation.xml.gz | 20.3 KB | Display | |
| Data in CIF |  emd_33265_validation.cif.gz emd_33265_validation.cif.gz | 26.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33265 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33265 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33265 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33265 | HTTPS FTP |
-Related structure data
| Related structure data |  7xkrMC  7xkhC  7xkoC  7xkpC  7xkqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33265.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33265.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.88 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_33265_msk_1.map emd_33265_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_33265_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_33265_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_33265_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : FoF1 from Bacillus PS3
| Entire | Name: FoF1 from Bacillus PS3 |
|---|---|
| Components |
|
-Supramolecule #1: FoF1 from Bacillus PS3
| Supramolecule | Name: FoF1 from Bacillus PS3 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 530 KDa |
-Macromolecule #1: ATP synthase subunit alpha
| Macromolecule | Name: ATP synthase subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO / EC number: H+-transporting two-sector ATPase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 54.848598 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSIRAEEISA LIKQQIENYE SQIQVSDVGT VIQVGDGIAR AHGLDNVMSG ELVEFANGVM GMALNLEENN VGIVILGPYT GIKEGDEVR RTGRIMEVPV GEALIGRVVN PLGQPVDGLG PVETTETRPI ESPAPGVMDR RSVHEPLQTG IKAIDALVPI G RGQRELII ...String: MSIRAEEISA LIKQQIENYE SQIQVSDVGT VIQVGDGIAR AHGLDNVMSG ELVEFANGVM GMALNLEENN VGIVILGPYT GIKEGDEVR RTGRIMEVPV GEALIGRVVN PLGQPVDGLG PVETTETRPI ESPAPGVMDR RSVHEPLQTG IKAIDALVPI G RGQRELII GDRQTGKTSV AIDTIINQKD QNMISIYVAI GQKESTVRTV VETLRKHGAL DYTIVVTASA SQPAPLLFLA PY AGVAMGE YFMYKGKHVL VVYDDLSKQA AAYRELSLLL RRPPGREAYP GDIFYLHSRL LERAAKLSDA KGGGSLTALP FVE TQAGDI SAYIPTNVIS ITDGQIFLQS DLFFSGVRPA INAGLSVSRV GGAAQIKAMK KVAGTLRLDL AAYRELEAFA QFGS DLDKA TQAKLARGAR TVEVLKQDLH QPIPVEKQVL IIYALTRGFL DDIPVEDVRR FEKEFYLFLD QNGQHLLEHI RTTKD LPNE DDLNKAIEAF KKTFVVSQ UniProtKB: ATP synthase subunit alpha |
-Macromolecule #2: ATP synthase subunit beta
| Macromolecule | Name: ATP synthase subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO / EC number: H+-transporting two-sector ATPase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 53.424625 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHHHH HMTRGRVIQV MGPVVDVKFE NGHLPAIYNA LKIQHKARNE NEVDIDLTLE VALHLGDDTV RTIAMASTDG LIRGMEVID TGAPISVPVG EVTLGRVFNV LGEPIDLEGD IPADARRDPI HRPAPKFEEL ATEVEILETG IKVVDLLAPY I KGGKIGLF ...String: MHHHHHHHHH HMTRGRVIQV MGPVVDVKFE NGHLPAIYNA LKIQHKARNE NEVDIDLTLE VALHLGDDTV RTIAMASTDG LIRGMEVID TGAPISVPVG EVTLGRVFNV LGEPIDLEGD IPADARRDPI HRPAPKFEEL ATEVEILETG IKVVDLLAPY I KGGKIGLF GGAGVGKTVL IQELIHNIAQ EHGGISVFAG VGERTREGND LYHEMKDSGV ISKTAMVFGQ MNEPPGARMR VA LTGLTMA EYFRDEQGQD VLLFIDNIFR FTQAGSEVSA LLGRMPSAVG YQPTLATEMG QLQERITSTA KGSITSIQAI YVP ADDYTD PAPATTFSHL DATTNLERKL AEMGIYPAVD PLASTSRALA PEIVGEEHYQ VARKVQQTLQ RYKELQDIIA ILGM DELSD EDKLVVHRAR RIQFFLSQNF HVAEQFTGQP GSYVPVKETV RGFKEILEGK YDHLPEDAFR LVGRIEEVVE KAKAM GVEV UniProtKB: ATP synthase subunit beta |
-Macromolecule #3: ATP synthase gamma chain
| Macromolecule | Name: ATP synthase gamma chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 31.859523 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASLRDIKTR INATKKTSQI TKAMEMVSTS KLNRAEQNAK SFVPYMEKIQ EVVANVALGA GGASHPMLVS RPVKKTGYLV ITSDRGLAG AYNSNVLRLV YQTIQKRHAS PDEYAIIVIG RVGLSFFRKR NMPVILDITR LPDQPSFADI KEIARKTVGL F ADGTFDEL ...String: MASLRDIKTR INATKKTSQI TKAMEMVSTS KLNRAEQNAK SFVPYMEKIQ EVVANVALGA GGASHPMLVS RPVKKTGYLV ITSDRGLAG AYNSNVLRLV YQTIQKRHAS PDEYAIIVIG RVGLSFFRKR NMPVILDITR LPDQPSFADI KEIARKTVGL F ADGTFDEL YMYYNHYVSA IQQEVTERKL LPLTDLAENK QRTVYEFEPS QEEILDVLLP QYAESLIYGA LLDAKASEHA AR MTAMKNA TDNANELIRT LTLSYNRARQ AAITQEITEI VAGANALQ UniProtKB: ATP synthase gamma chain |
-Macromolecule #4: ATP synthase epsilon chain
| Macromolecule | Name: ATP synthase epsilon chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.585905 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIHVSVVT PDGPVYEDDV EMVSVKAKSG ELGILPGHIP LVAPLEISAA RLKKGGKTQY IAVSGGFLEV RPDKVTILAQ AAERAEDID VLRAKAAKER AERRLQSQQD DIDFKRAELA LKRAMNRLSV AEMK UniProtKB: ATP synthase epsilon chain |
-Macromolecule #5: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 4 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 5 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #7: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 7 / Number of copies: 2 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: MOLYBDENUM / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 9625 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.0291 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)