[English] 日本語
 Yorodumi
Yorodumi- EMDB-33278: epsilon C-terminal domain deleted FoF1 from Bacillus PS3,state1,u... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | epsilon C-terminal domain deleted FoF1 from Bacillus PS3,state1,unisite condition | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | ATP synthase F1 ATPase FoF1 / MOTOR PROTEIN | |||||||||||||||
| Biological species |  | |||||||||||||||
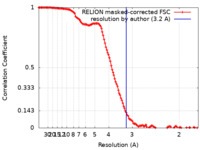
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||||||||
 Authors Authors | Nakano A / Kishikawa J / Nakanishi A / Mitsuoka K / Yokoyama K | |||||||||||||||
| Funding support |  Japan, 4 items Japan, 4 items
| |||||||||||||||
 Citation Citation |  Journal: PNAS Nexus / Year: 2022 Journal: PNAS Nexus / Year: 2022Title: Structural basis of unisite catalysis of bacterial FF-ATPase. Authors: Atsuki Nakano / Jun-Ichi Kishikawa / Atsuko Nakanishi / Kaoru Mitsuoka / Ken Yokoyama /  Abstract: Adenosine triphosphate (ATP) synthases (FF-ATPases) are crucial for all aerobic organisms. F, a water-soluble domain, can catalyze both the synthesis and hydrolysis of ATP with the rotation of the ...Adenosine triphosphate (ATP) synthases (FF-ATPases) are crucial for all aerobic organisms. F, a water-soluble domain, can catalyze both the synthesis and hydrolysis of ATP with the rotation of the central rotor inside a cylinder made of in three different conformations (referred to as , , and ). In this study, we determined multiple cryo-electron microscopy structures of bacterial FF exposed to different reaction conditions. The structures of nucleotide-depleted FF indicate that the ε subunit directly forces to adopt a closed form independent of the nucleotide binding to . The structure of FF under conditions that permit only a single catalytic subunit per enzyme to bind ATP is referred to as unisite catalysis and reveals that ATP hydrolysis unexpectedly occurs on instead of , where ATP hydrolysis proceeds in the steady-state catalysis of FF. This indicates that the unisite catalysis of bacterial FF significantly differs from the kinetics of steady-state turnover with continuous rotation of the shaft. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33278.map.gz emd_33278.map.gz | 140.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33278-v30.xml emd-33278-v30.xml emd-33278.xml emd-33278.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33278_fsc.xml emd_33278_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_33278.png emd_33278.png | 104.7 KB | ||
| Masks |  emd_33278_msk_1.map emd_33278_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-33278.cif.gz emd-33278.cif.gz | 4.1 KB | ||
| Others |  emd_33278_additional_1.map.gz emd_33278_additional_1.map.gz emd_33278_half_map_1.map.gz emd_33278_half_map_1.map.gz emd_33278_half_map_2.map.gz emd_33278_half_map_2.map.gz | 166.4 MB 140.8 MB 140.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33278 http://ftp.pdbj.org/pub/emdb/structures/EMD-33278 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33278 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33278 | HTTPS FTP |
-Validation report
| Summary document |  emd_33278_validation.pdf.gz emd_33278_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33278_full_validation.pdf.gz emd_33278_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_33278_validation.xml.gz emd_33278_validation.xml.gz | 20 KB | Display | |
| Data in CIF |  emd_33278_validation.cif.gz emd_33278_validation.cif.gz | 26.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33278 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33278 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33278 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33278 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33278.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33278.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.88 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_33278_msk_1.map emd_33278_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_33278_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_33278_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_33278_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : FoF1 from Bacillus sp. PS3
| Entire | Name: FoF1 from Bacillus sp. PS3 |
|---|---|
| Components |
|
-Supramolecule #1: FoF1 from Bacillus sp. PS3
| Supramolecule | Name: FoF1 from Bacillus sp. PS3 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 530 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: MOLYBDENUM / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 7329 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.001956 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|
 Movie
Movie Controller
Controller





























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)