+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Cryo-EM structure of a human pre-40S ribosomal subunit - State RRP12-A1 (with CK1) | |||||||||
 マップデータ マップデータ | ||||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | ribosome biogenesis / 40S ribosome / RIBOSOME | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報peptidyl-glutamine methylation / tRNA (m2G10) methyltransferase complex / rRNA (guanine-N7)-methylation / regulation of protein localization to nucleolus / tRNA methyltransferase activator activity / rRNA (guanine) methyltransferase activity / tRNA modification in the nucleus and cytosol / intermediate filament cytoskeleton organization / Activation of SMO / Methylation ...peptidyl-glutamine methylation / tRNA (m2G10) methyltransferase complex / rRNA (guanine-N7)-methylation / regulation of protein localization to nucleolus / tRNA methyltransferase activator activity / rRNA (guanine) methyltransferase activity / tRNA modification in the nucleus and cytosol / intermediate filament cytoskeleton organization / Activation of SMO / Methylation / trophectodermal cell differentiation / endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / negative regulation of NLRP3 inflammasome complex assembly / protein methyltransferase activity / tRNA methylation / cellular response to nutrient / positive regulation of rRNA processing / beta-catenin destruction complex / APC truncation mutants have impaired AXIN binding / AXIN missense mutants destabilize the destruction complex / Truncations of AMER1 destabilize the destruction complex / Beta-catenin phosphorylation cascade / Signaling by GSK3beta mutants / CTNNB1 S33 mutants aren't phosphorylated / CTNNB1 S37 mutants aren't phosphorylated / CTNNB1 S45 mutants aren't phosphorylated / CTNNB1 T41 mutants aren't phosphorylated / positive regulation of respiratory burst involved in inflammatory response / nucleolus organization / negative regulation of RNA splicing / rRNA methylation / Disassembly of the destruction complex and recruitment of AXIN to the membrane / neural crest cell differentiation / Maturation of nucleoprotein / U3 snoRNA binding / negative regulation of bicellular tight junction assembly / rRNA modification in the nucleus and cytosol / erythrocyte homeostasis / Formation of the ternary complex, and subsequently, the 43S complex / cytoplasmic side of rough endoplasmic reticulum membrane / snoRNA binding / preribosome, small subunit precursor / negative regulation of ubiquitin protein ligase activity / Ribosomal scanning and start codon recognition / Translation initiation complex formation / positive regulation of Rho protein signal transduction / Golgi organization / fibroblast growth factor binding / monocyte chemotaxis / TOR signaling / Protein hydroxylation / SARS-CoV-1 modulates host translation machinery / cellular response to ethanol / mTORC1-mediated signalling / Peptide chain elongation / Selenocysteine synthesis / Formation of a pool of free 40S subunits / positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator / Eukaryotic Translation Termination / ubiquitin ligase inhibitor activity / SRP-dependent cotranslational protein targeting to membrane / Response of EIF2AK4 (GCN2) to amino acid deficiency / negative regulation of ubiquitin-dependent protein catabolic process / positive regulation of signal transduction by p53 class mediator / Viral mRNA Translation / negative regulation of respiratory burst involved in inflammatory response / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / 90S preribosome / GTP hydrolysis and joining of the 60S ribosomal subunit / L13a-mediated translational silencing of Ceruloplasmin expression / Major pathway of rRNA processing in the nucleolus and cytosol / regulation of translational fidelity / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / maturation of LSU-rRNA / Nuclear events stimulated by ALK signaling in cancer / rough endoplasmic reticulum / translation regulator activity / ribosomal small subunit export from nucleus / positive regulation of cell cycle / translation initiation factor binding / Amplification of signal from unattached kinetochores via a MAD2 inhibitory signal / Maturation of protein E / Maturation of protein E / MDM2/MDM4 family protein binding / ER Quality Control Compartment (ERQC) / Myoclonic epilepsy of Lafora / FLT3 signaling by CBL mutants / IRAK2 mediated activation of TAK1 complex / Prevention of phagosomal-lysosomal fusion / Alpha-protein kinase 1 signaling pathway / Glycogen synthesis / IRAK1 recruits IKK complex / IRAK1 recruits IKK complex upon TLR7/8 or 9 stimulation / positive regulation of TORC1 signaling / liver regeneration / Endosomal Sorting Complex Required For Transport (ESCRT) / Membrane binding and targetting of GAG proteins / Negative regulation of FLT3 / Regulation of TBK1, IKKε (IKBKE)-mediated activation of IRF3, IRF7 / PTK6 Regulates RTKs and Their Effectors AKT1 and DOK1 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
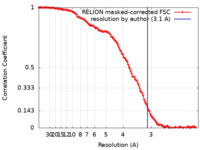
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.1 Å | |||||||||
 データ登録者 データ登録者 | Cheng J / Lau B / Thoms M / Ameismeier M / Berninghausen O / Hurt E / Beckmann R | |||||||||
| 資金援助 | 1件
| |||||||||
 引用 引用 |  ジャーナル: Nucleic Acids Res / 年: 2022 ジャーナル: Nucleic Acids Res / 年: 2022タイトル: The nucleoplasmic phase of pre-40S formation prior to nuclear export. 著者: Jingdong Cheng / Benjamin Lau / Matthias Thoms / Michael Ameismeier / Otto Berninghausen / Ed Hurt / Roland Beckmann /   要旨: Biogenesis of the small ribosomal subunit in eukaryotes starts in the nucleolus with the formation of a 90S precursor and ends in the cytoplasm. Here, we elucidate the enigmatic structural ...Biogenesis of the small ribosomal subunit in eukaryotes starts in the nucleolus with the formation of a 90S precursor and ends in the cytoplasm. Here, we elucidate the enigmatic structural transitions of assembly intermediates from human and yeast cells during the nucleoplasmic maturation phase. After dissociation of all 90S factors, the 40S body adopts a close-to-mature conformation, whereas the 3' major domain, later forming the 40S head, remains entirely immature. A first coordination is facilitated by the assembly factors TSR1 and BUD23-TRMT112, followed by re-positioning of RRP12 that is already recruited early to the 90S for further head rearrangements. Eventually, the uS2 cluster, CK1 (Hrr25 in yeast) and the export factor SLX9 associate with the pre-40S to provide export competence. These exemplary findings reveal the evolutionary conserved mechanism of how yeast and humans assemble the 40S ribosomal subunit, but reveal also a few minor differences. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_32800.map.gz emd_32800.map.gz | 106.2 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-32800-v30.xml emd-32800-v30.xml emd-32800.xml emd-32800.xml | 52.8 KB 52.8 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_32800_fsc.xml emd_32800_fsc.xml | 12.7 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_32800.png emd_32800.png | 163.9 KB | ||
| Filedesc metadata |  emd-32800.cif.gz emd-32800.cif.gz | 12.5 KB | ||
| その他 |  emd_32800_additional_1.map.gz emd_32800_additional_1.map.gz emd_32800_additional_2.map.gz emd_32800_additional_2.map.gz emd_32800_additional_3.map.gz emd_32800_additional_3.map.gz | 107.8 MB 107.8 MB 116.7 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32800 http://ftp.pdbj.org/pub/emdb/structures/EMD-32800 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32800 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32800 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_32800_validation.pdf.gz emd_32800_validation.pdf.gz | 458.7 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_32800_full_validation.pdf.gz emd_32800_full_validation.pdf.gz | 458.3 KB | 表示 | |
| XML形式データ |  emd_32800_validation.xml.gz emd_32800_validation.xml.gz | 13.1 KB | 表示 | |
| CIF形式データ |  emd_32800_validation.cif.gz emd_32800_validation.cif.gz | 17.7 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32800 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32800 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32800 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32800 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  7wttMC  7wtnC  7wtoC  7wtpC  7wtqC  7wtrC  7wtsC  7wtuC  7wtvC  7wtwC  7wtxC  7wtzC  7wu0C M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_32800.map.gz / 形式: CCP4 / 大きさ: 178 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_32800.map.gz / 形式: CCP4 / 大きさ: 178 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.059 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-追加マップ: #3
| ファイル | emd_32800_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-追加マップ: #2
| ファイル | emd_32800_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-追加マップ: #1
| ファイル | emd_32800_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
+全体 : Yeast pre-40S ribosomal subunit
+超分子 #1: Yeast pre-40S ribosomal subunit
+分子 #1: 18S rRNA
+分子 #2: 40S ribosomal protein S17
+分子 #3: 40S ribosomal protein S27
+分子 #4: 40S ribosomal protein S3a
+分子 #5: 40S ribosomal protein S28
+分子 #6: 40S ribosomal protein S4, X isoform
+分子 #7: 40S ribosomal protein S30
+分子 #8: 40S ribosomal protein S5
+分子 #9: 40S ribosomal protein S7
+分子 #10: 40S ribosomal protein S6
+分子 #11: 40S ribosomal protein S25
+分子 #12: 40S ribosomal protein S24
+分子 #13: RNA-binding protein PNO1
+分子 #14: 40S ribosomal protein S23
+分子 #15: Bystin
+分子 #16: 40S ribosomal protein S15a
+分子 #17: Pre-rRNA-processing protein TSR1 homolog
+分子 #18: Protein LTV1 homolog
+分子 #19: 40S ribosomal protein S19
+分子 #20: 40S ribosomal protein S18
+分子 #21: 40S ribosomal protein S16
+分子 #22: 40S ribosomal protein S15
+分子 #23: 40S ribosomal protein S14
+分子 #24: 40S ribosomal protein S13
+分子 #25: 40S ribosomal protein S11
+分子 #26: 40S ribosomal protein S9
+分子 #27: 40S ribosomal protein S8
+分子 #28: Multifunctional methyltransferase subunit TRM112-like protein
+分子 #29: Probable 18S rRNA (guanine-N(7))-methyltransferase
+分子 #30: RRP12-like protein
+分子 #31: 40S ribosomal protein S12
+分子 #32: Ubiquitin-40S ribosomal protein S27a
+分子 #33: Casein kinase I isoform alpha
+分子 #34: Ribosome biogenesis protein SLX9 homolog
+分子 #35: S-ADENOSYL-L-HOMOCYSTEINE
+分子 #36: ZINC ION
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.4 |
|---|---|
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 検出モード: COUNTING / 平均電子線量: 44.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 2.5 µm / 最小 デフォーカス(公称値): 0.8 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー






















































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)