[English] 日本語
 Yorodumi
Yorodumi- EMDB-32798: Cryo-EM structure of a yeast pre-40S ribosomal subunit - State Rio2-C -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a yeast pre-40S ribosomal subunit - State Rio2-C | |||||||||
 Map data Map data | local resolution filtered | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
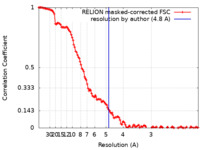
| Method | single particle reconstruction / cryo EM / Resolution: 4.8 Å | |||||||||
 Authors Authors | Cheng J / Lau B / Thoms M / Ameismeier M / Berninghausen O / Hurt E / Beckmann R | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2022 Journal: Nucleic Acids Res / Year: 2022Title: The nucleoplasmic phase of pre-40S formation prior to nuclear export. Authors: Jingdong Cheng / Benjamin Lau / Matthias Thoms / Michael Ameismeier / Otto Berninghausen / Ed Hurt / Roland Beckmann /   Abstract: Biogenesis of the small ribosomal subunit in eukaryotes starts in the nucleolus with the formation of a 90S precursor and ends in the cytoplasm. Here, we elucidate the enigmatic structural ...Biogenesis of the small ribosomal subunit in eukaryotes starts in the nucleolus with the formation of a 90S precursor and ends in the cytoplasm. Here, we elucidate the enigmatic structural transitions of assembly intermediates from human and yeast cells during the nucleoplasmic maturation phase. After dissociation of all 90S factors, the 40S body adopts a close-to-mature conformation, whereas the 3' major domain, later forming the 40S head, remains entirely immature. A first coordination is facilitated by the assembly factors TSR1 and BUD23-TRMT112, followed by re-positioning of RRP12 that is already recruited early to the 90S for further head rearrangements. Eventually, the uS2 cluster, CK1 (Hrr25 in yeast) and the export factor SLX9 associate with the pre-40S to provide export competence. These exemplary findings reveal the evolutionary conserved mechanism of how yeast and humans assemble the 40S ribosomal subunit, but reveal also a few minor differences. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32798.map.gz emd_32798.map.gz | 105.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32798-v30.xml emd-32798-v30.xml emd-32798.xml emd-32798.xml | 7.8 KB 7.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32798_fsc.xml emd_32798_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_32798.png emd_32798.png | 140.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32798 http://ftp.pdbj.org/pub/emdb/structures/EMD-32798 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32798 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32798 | HTTPS FTP |
-Related structure data
| Related structure data |  7wtnC  7wtoC  7wtpC  7wtqC  7wtrC  7wtsC  7wttC  7wtuC  7wtvC  7wtwC  7wtxC  7wtzC  7wu0C C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32798.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32798.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | local resolution filtered | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.059 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Human pre-40S ribosomal subunit
| Entire | Name: Human pre-40S ribosomal subunit |
|---|---|
| Components |
|
-Supramolecule #1: Human pre-40S ribosomal subunit
| Supramolecule | Name: Human pre-40S ribosomal subunit / type: complex / Chimera: Yes / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 44.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)