[English] 日本語
 Yorodumi
Yorodumi- EMDB-32768: Cryo-EM structure of the N-terminal deletion mutant of human pann... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-32768 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the N-terminal deletion mutant of human pannexin-1 in a nanodisc | ||||||||||||
 Map data Map data | 3D map of the N-terminal deletion mutant of human pannexin-1 | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | ATP release channel / vertebrate innexin homolog / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationATP transmembrane transporter activity / ATP transport / leak channel activity / Electric Transmission Across Gap Junctions / positive regulation of interleukin-1 alpha production / monoatomic anion transmembrane transport / bleb / wide pore channel activity / gap junction / monoatomic anion channel activity ...ATP transmembrane transporter activity / ATP transport / leak channel activity / Electric Transmission Across Gap Junctions / positive regulation of interleukin-1 alpha production / monoatomic anion transmembrane transport / bleb / wide pore channel activity / gap junction / monoatomic anion channel activity / gap junction channel activity / positive regulation of macrophage cytokine production / Mechanical load activates signaling by PIEZO1 and integrins in osteocytes / oogenesis / response to ATP / The NLRP3 inflammasome / monoatomic cation transport / response to ischemia / positive regulation of interleukin-1 beta production / calcium channel activity / actin filament binding / calcium ion transport / cell-cell signaling / protease binding / scaffold protein binding / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / transmembrane transporter binding / signaling receptor binding / endoplasmic reticulum membrane / structural molecule activity / endoplasmic reticulum / protein-containing complex / identical protein binding / membrane / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
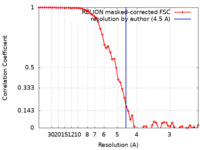
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | ||||||||||||
 Authors Authors | Kuzuya M / Hirano H | ||||||||||||
| Funding support |  Japan, 3 items Japan, 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Signal / Year: 2022 Journal: Sci Signal / Year: 2022Title: Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids. Authors: Maki Kuzuya / Hidemi Hirano / Kenichi Hayashida / Masakatsu Watanabe / Kazumi Kobayashi / Tohru Terada / Md Iqbal Mahmood / Florence Tama / Kazutoshi Tani / Yoshinori Fujiyoshi / Atsunori Oshima /  Abstract: Pannexin (PANX) family proteins form large-pore channels that mediate purinergic signaling. We analyzed the cryo-EM structures of human PANX1 in lipid nanodiscs to elucidate the gating mechanism and ...Pannexin (PANX) family proteins form large-pore channels that mediate purinergic signaling. We analyzed the cryo-EM structures of human PANX1 in lipid nanodiscs to elucidate the gating mechanism and its regulation by the amino terminus in phospholipids. The wild-type channel has an amino-terminal funnel in the pore, but in the presence of the inhibitor probenecid, a cytoplasmically oriented amino terminus and phospholipids obstruct the pore. Functional analysis using whole-cell patch-clamp and oocyte voltage clamp showed that PANX1 lacking the amino terminus did not open and had a dominant negative effect on channel activity, thus confirming that the amino-terminal domain played an essential role in channel opening. These observations suggest that dynamic conformational changes in the amino terminus of human PANX1 are associated with lipid movement in and out of the pore. Moreover, the data provide insight into the gating mechanism of PANX1 and, more broadly, other large-pore channels. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32768.map.gz emd_32768.map.gz | 14.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32768-v30.xml emd-32768-v30.xml emd-32768.xml emd-32768.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32768_fsc.xml emd_32768_fsc.xml | 5.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_32768.png emd_32768.png | 223.6 KB | ||
| Masks |  emd_32768_msk_1.map emd_32768_msk_1.map | 15.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-32768.cif.gz emd-32768.cif.gz | 6.1 KB | ||
| Others |  emd_32768_half_map_1.map.gz emd_32768_half_map_1.map.gz emd_32768_half_map_2.map.gz emd_32768_half_map_2.map.gz | 11.9 MB 11.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32768 http://ftp.pdbj.org/pub/emdb/structures/EMD-32768 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32768 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32768 | HTTPS FTP |
-Related structure data
| Related structure data |  7wsvMC  7f8jC  7f8nC  7f8oC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32768.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32768.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D map of the N-terminal deletion mutant of human pannexin-1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.232 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
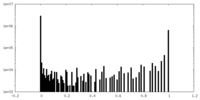
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_32768_msk_1.map emd_32768_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
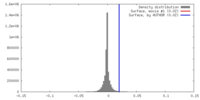
| Density Histograms |
-Half map: #1
| File | emd_32768_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
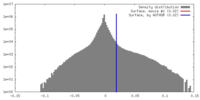
| Density Histograms |
-Half map: #2
| File | emd_32768_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
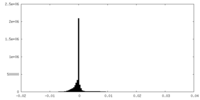
| Density Histograms |
- Sample components
Sample components
-Entire : Heptamar of N-terminal deleted human pannexin-1 in a nanodisc
| Entire | Name: Heptamar of N-terminal deleted human pannexin-1 in a nanodisc |
|---|---|
| Components |
|
-Supramolecule #1: Heptamar of N-terminal deleted human pannexin-1 in a nanodisc
| Supramolecule | Name: Heptamar of N-terminal deleted human pannexin-1 in a nanodisc type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Pannexin-1
| Macromolecule | Name: Pannexin-1 / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.955438 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MTEPKFKGLR LELAVDKMVT CIAVGLPLLL ISLAFAQEIS IGTQISCFSP SSFSWRQAAF VDSYCWAAVQ QKNSLQSESG NLPLWLHKF FPYILLLFAI LLYLPPLFWR FAAAPHICSD LKFIMEELDK VYNRAIKAAK SARDLDMRDG ACSVPGVTEN L GQSLWEVS ...String: MTEPKFKGLR LELAVDKMVT CIAVGLPLLL ISLAFAQEIS IGTQISCFSP SSFSWRQAAF VDSYCWAAVQ QKNSLQSESG NLPLWLHKF FPYILLLFAI LLYLPPLFWR FAAAPHICSD LKFIMEELDK VYNRAIKAAK SARDLDMRDG ACSVPGVTEN L GQSLWEVS ESHFKYPIVE QYLKTKKNSN NLIIKYISCR LLTLIIILLA CIYLGYYFSL SSLSDEFVCS IKSGILRNDS TV PDQFQCK LIAVGIFQLL SVINLVVYVL LAPVVVYTLF VPFRQKTDVL KVYEILPTFD VLHFKSEGYN DLSLYNLFLE ENI SEVKSY KCLKVLENIK SSGQGIDPML LLTNLGMIKM DVVDGKTPMS AEMREEQGNQ TAELQGMNID SETKANNGEK NARQ RLLDS SC UniProtKB: Pannexin-1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.1 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: pH 7.5 was used. | |||||||||
| Grid | Model: Quantifoil R2/2 / Material: MOLYBDENUM / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | |||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA KF80 Details: Blot for 10 seconds at room temperature followed by plunge freezing. Humidity and temperature are not controlled.. |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3000SFF |
|---|---|
| Temperature | Min: 80.0 K / Max: 100.0 K |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Number grids imaged: 1 / Number real images: 3587 / Average exposure time: 8.0 sec. / Average electron dose: 56.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 3.5 µm / Calibrated defocus min: 1.4000000000000001 µm / Calibrated magnification: 40600 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 1.6 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.4000000000000001 µm / Nominal magnification: 30000 |
| Sample stage | Specimen holder model: JEOL / Cooling holder cryogen: HELIUM |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 344.6 |
|---|---|
| Output model |  PDB-7wsv: |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)