+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21150 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Xenopus tropicalis pannexin 1 channel | |||||||||
 Map data Map data | Sharpened map of a pannexin1 channel | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | channel / ATP release / heptamer / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationwide pore channel activity / positive regulation of interleukin-1 production / gap junction / monoatomic cation transport / cell-cell signaling / monoatomic ion transmembrane transport / endoplasmic reticulum membrane / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species | ||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.02 Å | |||||||||
 Authors Authors | Syrjanen JL / Furukawa H | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: The Cryo-EM structure of pannexin 1 reveals unique motifs for ion selection and inhibition. Authors: Kevin Michalski / Johanna L Syrjanen / Erik Henze / Julia Kumpf / Hiro Furukawa / Toshimitsu Kawate /  Abstract: Pannexins are large-pore forming channels responsible for ATP release under a variety of physiological and pathological conditions. Although predicted to share similar membrane topology with other ...Pannexins are large-pore forming channels responsible for ATP release under a variety of physiological and pathological conditions. Although predicted to share similar membrane topology with other large-pore forming proteins such as connexins, innexins, and LRRC8, pannexins have minimal sequence similarity to these protein families. Here, we present the cryo-EM structure of a frog pannexin 1 (Panx1) channel at 3.0 Å. We find that Panx1 protomers harbor four transmembrane helices similar in arrangement to other large-pore forming proteins but assemble as a heptameric channel with a unique constriction formed by Trp74 in the first extracellular loop. Mutating Trp74 or the nearby Arg75 disrupt ion selectivity, whereas altering residues in the hydrophobic groove formed by the two extracellular loops abrogates channel inhibition by carbenoxolone. Our structural and functional study establishes the extracellular loops as important structural motifs for ion selectivity and channel inhibition in Panx1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21150.map.gz emd_21150.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21150-v30.xml emd-21150-v30.xml emd-21150.xml emd-21150.xml | 11.9 KB 11.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_21150.png emd_21150.png | 153.5 KB | ||
| Filedesc metadata |  emd-21150.cif.gz emd-21150.cif.gz | 5.2 KB | ||
| Others |  emd_21150_additional.map.gz emd_21150_additional.map.gz | 59.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21150 http://ftp.pdbj.org/pub/emdb/structures/EMD-21150 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21150 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21150 | HTTPS FTP |
-Related structure data
| Related structure data |  6vd7MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_21150.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21150.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map of a pannexin1 channel | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
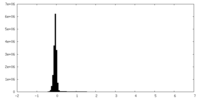
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: unsharpened map of a pannexin1 channel
| File | emd_21150_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map of a pannexin1 channel | ||||||||||||
| Projections & Slices |
| ||||||||||||
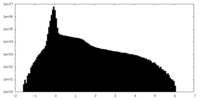
| Density Histograms |
- Sample components
Sample components
-Entire : Heptameric pannexin1 complex assembly
| Entire | Name: Heptameric pannexin1 complex assembly |
|---|---|
| Components |
|
-Supramolecule #1: Heptameric pannexin1 complex assembly
| Supramolecule | Name: Heptameric pannexin1 complex assembly / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism: |
-Macromolecule #1: Pannexin
| Macromolecule | Name: Pannexin / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 39.424148 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAIAHIATEY VFSDFLLKDP PESKYKGLRL ELAVDKLVSC IAVGLPLLLI SLAFAQEITL GSQISCFAPT SFSWRQAAYV DSFCWAAVQ QKHLSQSDSG NVPLWLHKFF PYILLLVAVL LYLPNLFWRF TAAPHLSSDL KFVMEELDKC YNRDIKDIKA A NNLNSSDK ...String: MAIAHIATEY VFSDFLLKDP PESKYKGLRL ELAVDKLVSC IAVGLPLLLI SLAFAQEITL GSQISCFAPT SFSWRQAAYV DSFCWAAVQ QKHLSQSDSG NVPLWLHKFF PYILLLVAVL LYLPNLFWRF TAAPHLSSDL KFVMEELDKC YNRDIKDIKA A NNLNSSDK RDYPIVEQYL KTKNNSYGLI IKYLICRVVT LIIVFTACIY LGYYISLFSL TDEFTCNIRT GILRNDTALP PL VQCKLIA VGVFRLLSYI NLIIYVLIMP FIIYAMLVPF RKTANVLKVY EVLPTFSVQQ APSKTYDDHS LFLLFLEENV SEL KSYKFL KVLENIKASS WSHPQFEK UniProtKB: Pannexin |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Material: COPPER / Support film - Material: CARBON / Support film - topology: LACEY / Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Chamber temperature: 288.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 57.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C7 (7 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 3.02 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cisTEM / Number images used: 90185 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)