+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-32465 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Subcomplexes A and E in NDH complex from Arabidopsis | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Subcomplex A / subcomplex E / supercomplex / Arabidopsis / plant / cyclic electron transport / ELECTRON TRANSPORT | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationNADH dehydrogenase complex (plastoquinone) assembly / NAD(P)H dehydrogenase complex (plastoquinone) / cellular response to sulfate starvation / thylakoid membrane / chloroplast thylakoid / NADPH dehydrogenase activity / Translocases; Catalysing the translocation of protons; Linked to oxidoreductase reactions / chloroplast envelope / photosynthetic electron transport in photosystem I / oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor ...NADH dehydrogenase complex (plastoquinone) assembly / NAD(P)H dehydrogenase complex (plastoquinone) / cellular response to sulfate starvation / thylakoid membrane / chloroplast thylakoid / NADPH dehydrogenase activity / Translocases; Catalysing the translocation of protons; Linked to oxidoreductase reactions / chloroplast envelope / photosynthetic electron transport in photosystem I / oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor / chloroplast stroma / plastid / photosynthesis, light reaction / chloroplast thylakoid membrane / NADH dehydrogenase (ubiquinone) activity / defense response to fungus / quinone binding / photosynthesis / chloroplast / NAD binding / 4 iron, 4 sulfur cluster binding / iron ion binding / plasma membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.33 Å | ||||||||||||||||||
 Authors Authors | Pan XW / Li M | ||||||||||||||||||
| Funding support |  China, 5 items China, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Mol Plant / Year: 2022 Journal: Mol Plant / Year: 2022Title: Supramolecular assembly of chloroplast NADH dehydrogenase-like complex with photosystem I from Arabidopsis thaliana. Authors: Xiaodong Su / Duanfang Cao / Xiaowei Pan / Lifang Shi / Zhenfeng Liu / Luca Dall'Osto / Roberto Bassi / Xinzheng Zhang / Mei Li /   Abstract: Cyclic electron transport/flow (CET/CEF) in chloroplasts is a regulatory process essential for the optimization of plant photosynthetic efficiency. A crucial CEF pathway is catalyzed by a membrane- ...Cyclic electron transport/flow (CET/CEF) in chloroplasts is a regulatory process essential for the optimization of plant photosynthetic efficiency. A crucial CEF pathway is catalyzed by a membrane-embedded NADH dehydrogenase-like (NDH) complex that contains at least 29 protein subunits and associates with photosystem I (PSI) to form the NDH-PSI supercomplex. Here, we report the 3.9 Å resolution structure of the Arabidopsis thaliana NDH-PSI (AtNDH-PSI) supercomplex. We constructed structural models for 26 AtNDH subunits, among which 11 are unique to chloroplasts and stabilize the core part of the NDH complex. In the supercomplex, one NDH can bind up to two PSI-light-harvesting complex I (PSI-LHCI) complexes at both sides of its membrane arm. Two minor LHCIs, Lhca5 and Lhca6, each present in one PSI-LHCI, interact with NDH and contribute to supercomplex formation and stabilization. Collectively, our study reveals the structural details of the AtNDH-PSI supercomplex assembly and provides a molecular basis for further investigation of the regulatory mechanism of CEF in plants. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32465.map.gz emd_32465.map.gz | 226.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32465-v30.xml emd-32465-v30.xml emd-32465.xml emd-32465.xml | 22.2 KB 22.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32465.png emd_32465.png | 38.1 KB | ||
| Filedesc metadata |  emd-32465.cif.gz emd-32465.cif.gz | 7.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32465 http://ftp.pdbj.org/pub/emdb/structures/EMD-32465 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32465 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32465 | HTTPS FTP |
-Validation report
| Summary document |  emd_32465_validation.pdf.gz emd_32465_validation.pdf.gz | 569.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32465_full_validation.pdf.gz emd_32465_full_validation.pdf.gz | 569.4 KB | Display | |
| Data in XML |  emd_32465_validation.xml.gz emd_32465_validation.xml.gz | 6.7 KB | Display | |
| Data in CIF |  emd_32465_validation.cif.gz emd_32465_validation.cif.gz | 7.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32465 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32465 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32465 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32465 | HTTPS FTP |
-Related structure data
| Related structure data |  7wfgMC  7wfdC  7wfeC  7wffC  7wg5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32465.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32465.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
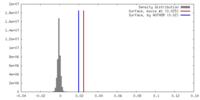
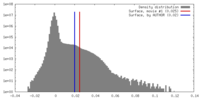
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Subcomplex A and E in NDH complex from Arabidopsis
+Supramolecule #1: Subcomplex A and E in NDH complex from Arabidopsis
+Macromolecule #1: NAD(P)H-quinone oxidoreductase subunit H, chloroplastic
+Macromolecule #2: NAD(P)H-quinone oxidoreductase subunit I, chloroplastic
+Macromolecule #3: NAD(P)H-quinone oxidoreductase subunit J, chloroplastic
+Macromolecule #4: NAD(P)H-quinone oxidoreductase subunit K, chloroplastic
+Macromolecule #5: NAD(P)H-quinone oxidoreductase subunit L, chloroplastic
+Macromolecule #6: NAD(P)H-quinone oxidoreductase subunit M, chloroplastic
+Macromolecule #7: NAD(P)H-quinone oxidoreductase subunit N, chloroplastic
+Macromolecule #8: NdhO
+Macromolecule #9: NdhT
+Macromolecule #10: IRON/SULFUR CLUSTER
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



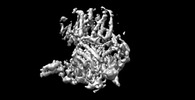




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)