+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
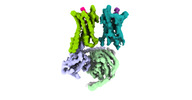
| Title | Cryo-EM structure of a dimeric GPCR-Gi complex with peptide | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / GPCR / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmesendoderm migration / mesoderm migration involved in gastrulation / cell migration involved in mesendoderm migration / apelin receptor binding / : / apelin receptor activity / apelin receptor signaling pathway / mechanoreceptor activity / regulation of gap junction assembly / positive regulation of G protein-coupled receptor internalization ...mesendoderm migration / mesoderm migration involved in gastrulation / cell migration involved in mesendoderm migration / apelin receptor binding / : / apelin receptor activity / apelin receptor signaling pathway / mechanoreceptor activity / regulation of gap junction assembly / positive regulation of G protein-coupled receptor internalization / vascular associated smooth muscle cell differentiation / atrioventricular valve development / regulation of body fluid levels / venous blood vessel development / positive regulation of heart contraction / positive regulation of cardiac muscle hypertrophy in response to stress / positive regulation of trophoblast cell migration / endocardial cushion formation / positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis / placenta blood vessel development / endoderm development / adult heart development / coronary vasculature development / vasculature development / embryonic heart tube development / G protein-coupled peptide receptor activity / aorta development / negative regulation of cardiac muscle hypertrophy in response to stress / ventricular septum morphogenesis / blood vessel development / heart looping / vasculogenesis / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / D2 dopamine receptor binding / response to prostaglandin E / gastrulation / adenylate cyclase regulator activity / G protein-coupled serotonin receptor binding / adenylate cyclase-inhibiting serotonin receptor signaling pathway / cellular response to forskolin / Peptide ligand-binding receptors / regulation of mitotic spindle organization / positive regulation of release of sequestered calcium ion into cytosol / Regulation of insulin secretion / positive regulation of cholesterol biosynthetic process / electron transport chain / hormone activity / negative regulation of insulin secretion / G protein-coupled receptor binding / response to peptide hormone / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / positive regulation of angiogenesis / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / GDP binding / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / signaling receptor activity / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / heart development Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.21 Å | |||||||||
 Authors Authors | Xu F / Yue Y | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: Structural insight into apelin receptor-G protein stoichiometry. Authors: Yang Yue / Lier Liu / Li-Jie Wu / Yiran Wu / Ling Wang / Fei Li / Junlin Liu / Gye-Won Han / Bo Chen / Xi Lin / Rebecca L Brouillette / Émile Breault / Jean-Michel Longpré / Songting Shi / ...Authors: Yang Yue / Lier Liu / Li-Jie Wu / Yiran Wu / Ling Wang / Fei Li / Junlin Liu / Gye-Won Han / Bo Chen / Xi Lin / Rebecca L Brouillette / Émile Breault / Jean-Michel Longpré / Songting Shi / Hui Lei / Philippe Sarret / Raymond C Stevens / Michael A Hanson / Fei Xu /    Abstract: The technique of cryogenic-electron microscopy (cryo-EM) has revolutionized the field of membrane protein structure and function with a focus on the dominantly observed molecular species. This report ...The technique of cryogenic-electron microscopy (cryo-EM) has revolutionized the field of membrane protein structure and function with a focus on the dominantly observed molecular species. This report describes the structural characterization of a fully active human apelin receptor (APJR) complexed with heterotrimeric G protein observed in both 2:1 and 1:1 stoichiometric ratios. We use cryo-EM single-particle analysis to determine the structural details of both species from the same sample preparation. Protein preparations, in the presence of the endogenous peptide ligand ELA or a synthetic small molecule, both demonstrate these mixed stoichiometric states. Structural differences in G protein engagement between dimeric and monomeric APJR suggest a role for the stoichiometry of G protein-coupled receptor- (GPCR-)G protein coupling on downstream signaling and receptor pharmacology. Furthermore, a small, hydrophobic dimer interface provides a starting framework for additional class A GPCR dimerization studies. Together, these findings uncover a mechanism of versatile regulation through oligomerization by which GPCRs can modulate their signaling. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32245.map.gz emd_32245.map.gz | 24.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32245-v30.xml emd-32245-v30.xml emd-32245.xml emd-32245.xml | 21.9 KB 21.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32245.png emd_32245.png | 35.3 KB | ||
| Filedesc metadata |  emd-32245.cif.gz emd-32245.cif.gz | 6.5 KB | ||
| Others |  emd_32245_additional_1.map.gz emd_32245_additional_1.map.gz emd_32245_half_map_1.map.gz emd_32245_half_map_1.map.gz emd_32245_half_map_2.map.gz emd_32245_half_map_2.map.gz | 13.5 MB 25 MB 25 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32245 http://ftp.pdbj.org/pub/emdb/structures/EMD-32245 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32245 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32245 | HTTPS FTP |
-Validation report
| Summary document |  emd_32245_validation.pdf.gz emd_32245_validation.pdf.gz | 710 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32245_full_validation.pdf.gz emd_32245_full_validation.pdf.gz | 709.5 KB | Display | |
| Data in XML |  emd_32245_validation.xml.gz emd_32245_validation.xml.gz | 10.8 KB | Display | |
| Data in CIF |  emd_32245_validation.cif.gz emd_32245_validation.cif.gz | 12.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32245 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32245 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32245 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32245 | HTTPS FTP |
-Related structure data
| Related structure data |  7w0nMC  7susC  7w0lC  7w0mC  7w0oC  7w0pC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32245.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32245.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.46667 Å | ||||||||||||||||||||||||||||||||||||
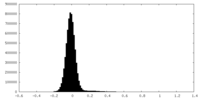
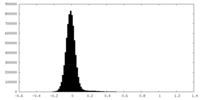
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_32245_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
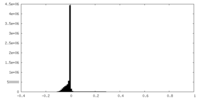
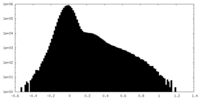
| Density Histograms |
-Half map: #2
| File | emd_32245_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
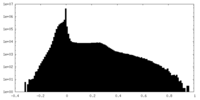
| Density Histograms |
-Half map: #1
| File | emd_32245_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
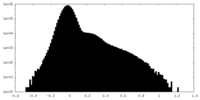
| Density Histograms |
- Sample components
Sample components
-Entire : GPCR-Gi Complex
| Entire | Name: GPCR-Gi Complex |
|---|---|
| Components |
|
-Supramolecule #1: GPCR-Gi Complex
| Supramolecule | Name: GPCR-Gi Complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.55916 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GSMGCTLSAE DKAAVERSKM IDRNLREDGE KAAREVKLLL LGAGESGKST IVKQMKIIHE AGYSEEECKQ YKAVVYSNTI QSIIAIIRA MGRLKIDFGD SARADDARQL FVLAGAAEEG FMTAELAGVI KRLWKDSGVQ ACFNRSREYQ LNDSAAYYLN D LDRIAQPN ...String: GSMGCTLSAE DKAAVERSKM IDRNLREDGE KAAREVKLLL LGAGESGKST IVKQMKIIHE AGYSEEECKQ YKAVVYSNTI QSIIAIIRA MGRLKIDFGD SARADDARQL FVLAGAAEEG FMTAELAGVI KRLWKDSGVQ ACFNRSREYQ LNDSAAYYLN D LDRIAQPN YIPTQQDVLR TRVKTTGIVE THFTFKDLHF KMFDVGGQRS ERKKWIHCFE GVTAIIFCVA LSDYDLVLAE DE EMNRMHE SMKLFDSICN NKWFTDTSII LFLNKKDLFE EKIKKSPLTI CYPEYAGSNT YEEAAAYIQC QFEDLNKRKD TKE IYTHFT CATDTKNVQF VFDAVTDVII KNNLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.41693 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL ...String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD INAICFFPNG NA FATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAGHDNRVSC LGV TDDGMA VATGSWDSFL KIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Soluble cytochrome b562,Apelin receptor
| Macromolecule | Name: Soluble cytochrome b562,Apelin receptor / type: protein_or_peptide / ID: 4 Details: Fusion protein of Soluble cytochrome b562, linker and Apelin receptor Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 54.312578 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDKHHHHHH HHHHLEVLFQ GPADLEDNWE TLNDNLKVIE KADNAAQVKD ALTKMRAAAL DAQKATPPK LEDKSPDSPE MKDFRHGFDI LVGQIDDALK LANEGKVKEA QAAAEQLKTT RNAYIQKYLE EGGDFDNYYG A DNQSECEY ...String: MKTIIALSYI FCLVFADYKD DDDKHHHHHH HHHHLEVLFQ GPADLEDNWE TLNDNLKVIE KADNAAQVKD ALTKMRAAAL DAQKATPPK LEDKSPDSPE MKDFRHGFDI LVGQIDDALK LANEGKVKEA QAAAEQLKTT RNAYIQKYLE EGGDFDNYYG A DNQSECEY TDWKSSGALI PAIYMLVFLL GTTGNGLVLW TVFRSSREKR RSADIFIASL AVADLTFVVT LPLWATYTYR DY DWPFGTF FCKLSSYLIF VNMYASVFCL TGLSFDRYLA IVRPVANARL RLRVSGAVAT AVLWVLAALL AMPVMVLRTT GDL ENTTKV QCYMDYSMVA TVSSEWAWEV GLGVSSTTVG FVVPFTIMLT CYFFIAQTIA GHFRKERIEG LRKRRRLLSI IVVL VVTFA LCWMPYHLVK TLYMLGSLLH WPCDFDLFLM NIFPYCTCIS YVNSCLNPFL YAFFDPRFRQ ACTSMLCCGQ SR UniProtKB: Soluble cytochrome b562, Apelin receptor |
-Macromolecule #5: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 31.870629 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MLLVNQSHQG FNKEHTSKMV SAIVLYVLLA AAAHSAFADV QLVESGGGLV QPGGSRKLSC SASGFAFSSF GMHWVRQAPE KGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSEDT AMYYCVRSIY YYGSSPFDFW GQGTTLTVSS G GGGSGGGG ...String: MLLVNQSHQG FNKEHTSKMV SAIVLYVLLA AAAHSAFADV QLVESGGGLV QPGGSRKLSC SASGFAFSSF GMHWVRQAPE KGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSEDT AMYYCVRSIY YYGSSPFDFW GQGTTLTVSS G GGGSGGGG SGGGGSDIVM TQATSSVPVT PGESVSISCR SSKSLLHSNG NTYLYWFLQR PGQSPQLLIY RMSNLASGVP DR FSGSGSG TAFTLTISRL EAEDVGVYYC MQHLEYPLTF GAGTKLELKA AAHHHHHHHH |
-Macromolecule #6: Apelin receptor early endogenous ligand
| Macromolecule | Name: Apelin receptor early endogenous ligand / type: protein_or_peptide / ID: 6 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 3.980873 KDa |
| Sequence | String: QRPVNLTMRR KLRKHNCLQR RCMPLHSRVP FP UniProtKB: Apelin receptor early endogenous ligand |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.21 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 76513 |
| Initial angle assignment | Type: COMMON LINE |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller








































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)