[English] 日本語
 Yorodumi
Yorodumi- EMDB-32086: The malate-bound AtALMT1 structure at pH 7.5 (ALMT1malate/pH7.5) -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-32086 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | The malate-bound AtALMT1 structure at pH 7.5 (ALMT1malate/pH7.5) | |||||||||||||||
 Map data Map data | the malate-bound AtALMT1 structure at pH 7.5 (ALMT1malate/pH7.5) | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | ALMT1 / Aluminum Resistance / malate transport / TRANSPORT PROTEIN | |||||||||||||||
| Function / homology | malate transmembrane transport / malate transmembrane transporter activity / Aluminum-activated malate transporter / Aluminium activated malate transporter / response to aluminum ion / monoatomic ion transmembrane transport / plasma membrane / Aluminum-activated malate transporter 1 Function and homology information Function and homology information | |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||||||||
 Authors Authors | Wang JQ | |||||||||||||||
| Funding support |  China, 4 items China, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Cell Res / Year: 2022 Journal: Cell Res / Year: 2022Title: Structural basis of ALMT1-mediated aluminum resistance in Arabidopsis. Authors: Jiangqin Wang / Xiafei Yu / Zhong Jie Ding / Xiaokang Zhang / Yanping Luo / Ximing Xu / Yuan Xie / Xiaoxiao Li / Tian Yuan / Shao Jian Zheng / Wei Yang / Jiangtao Guo /  Abstract: The plant aluminum (Al)-activated malate transporter ALMT1 mediates the efflux of malate to chelate the Al in acidic soils and underlies the plant Al resistance. Here we present cryo-electron ...The plant aluminum (Al)-activated malate transporter ALMT1 mediates the efflux of malate to chelate the Al in acidic soils and underlies the plant Al resistance. Here we present cryo-electron microscopy (cryo-EM) structures of Arabidopsis thaliana ALMT1 (AtALMT1) in the apo, malate-bound, and Al-bound states at neutral and/or acidic pH at up to 3.0 Å resolution. The AtALMT1 dimer assembles an anion channel and each subunit contains six transmembrane helices (TMs) and six cytosolic α-helices. Two pairs of Arg residues are located in the center of the channel pore and contribute to malate recognition. Al binds at the extracellular side of AtALMT1 and induces conformational changes of the TM1-2 loop and the TM5-6 loop, resulting in the opening of the extracellular gate. These structures, along with electrophysiological measurements, molecular dynamic simulations, and mutagenesis study in Arabidopsis, elucidate the structural basis for Al-activated malate transport by ALMT1. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32086.map.gz emd_32086.map.gz | 20.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32086-v30.xml emd-32086-v30.xml emd-32086.xml emd-32086.xml | 13.7 KB 13.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32086.png emd_32086.png | 37.8 KB | ||
| Filedesc metadata |  emd-32086.cif.gz emd-32086.cif.gz | 5.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32086 http://ftp.pdbj.org/pub/emdb/structures/EMD-32086 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32086 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32086 | HTTPS FTP |
-Validation report
| Summary document |  emd_32086_validation.pdf.gz emd_32086_validation.pdf.gz | 527.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32086_full_validation.pdf.gz emd_32086_full_validation.pdf.gz | 527 KB | Display | |
| Data in XML |  emd_32086_validation.xml.gz emd_32086_validation.xml.gz | 5.6 KB | Display | |
| Data in CIF |  emd_32086_validation.cif.gz emd_32086_validation.cif.gz | 6.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32086 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32086 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32086 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32086 | HTTPS FTP |
-Related structure data
| Related structure data |  7vq5MC  7vojC  7vq3C  7vq4C  7vq7C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32086.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32086.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | the malate-bound AtALMT1 structure at pH 7.5 (ALMT1malate/pH7.5) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.014 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : the malate-bound AtALMT1 structure at pH 7.5 (ALMT1malate/pH7.5)
| Entire | Name: the malate-bound AtALMT1 structure at pH 7.5 (ALMT1malate/pH7.5) |
|---|---|
| Components |
|
-Supramolecule #1: the malate-bound AtALMT1 structure at pH 7.5 (ALMT1malate/pH7.5)
| Supramolecule | Name: the malate-bound AtALMT1 structure at pH 7.5 (ALMT1malate/pH7.5) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Aluminum-activated malate transporter 1
| Macromolecule | Name: Aluminum-activated malate transporter 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 56.833668 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEKVREIVRE GIRVGNEDPR RIIHAFKVGL ALVLVSSFYY YQPFGPFTDY FGINAMWAVM TVVVVFEFSV GATLGKGLNR GVATLVAGG LGIGAHQLAR LSGATVEPIL LVMLVFVQAA LSTFVRFFPW VKTKFDYGIL IFILTFALIS LSGFRDEEIM D LAESRLST ...String: MEKVREIVRE GIRVGNEDPR RIIHAFKVGL ALVLVSSFYY YQPFGPFTDY FGINAMWAVM TVVVVFEFSV GATLGKGLNR GVATLVAGG LGIGAHQLAR LSGATVEPIL LVMLVFVQAA LSTFVRFFPW VKTKFDYGIL IFILTFALIS LSGFRDEEIM D LAESRLST VVIGGVSCIL ISIFVCPVWA GQDLHSLLAS NFDTLSHFLQ DFGDEYFEAR EKGDYKVVEK RKKNLERYKS VL DSKSDEE ALANYAEWEP PHGQFRFRHP WKQYVAVGAL LRQCAYRIDA LNSYINSDFQ IPVDIKKKLE TPLRRMSSES GNS MKEMSI SLKQMIKSSS SDIHVSNSQA ACKSLSTLLK SGILNDVEPL QMISLMTTVS MLIDIVNLTE KISESVHELA SAAR FKNKM RPTVLYEKSD SGSIGRAMPI DSHEDHHVVT VLHDVDNDRS NNVDDSRGGS SQDSCHHVAI KIVDDNSNHE KHEDG EIHV HTLSNGHLQL EGGSSGGWSH PQFEK UniProtKB: Aluminum-activated malate transporter 1 |
-Macromolecule #2: (2S)-2-hydroxybutanedioic acid
| Macromolecule | Name: (2S)-2-hydroxybutanedioic acid / type: ligand / ID: 2 / Number of copies: 1 / Formula: LMR |
|---|---|
| Molecular weight | Theoretical: 134.087 Da |
| Chemical component information | 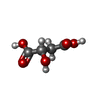 ChemComp-LMR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
| Details | For the sample of AtALMT1 bound to malate (ALMT1malate/pH7.5), the peak fractions after SEC were mixed with malate to a final concentration of 10 mM |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average exposure time: 8.0 sec. / Average electron dose: 64.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















