+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-32068 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human ATP13A2 (SPM-bound E2P state) | ||||||||||||
 Map data Map data | postprocess_masked.mrc from Relion3.1 | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | ATP13A2 / PARK9 / P-type ATPase / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationpolyamine transmembrane transport / spermine transmembrane transport / : / polyamine transmembrane transporter activity / ABC-type polyamine transporter activity / regulation of autophagosome size / extracellular exosome biogenesis / negative regulation of lysosomal protein catabolic process / regulation of chaperone-mediated autophagy / P-type ion transporter activity ...polyamine transmembrane transport / spermine transmembrane transport / : / polyamine transmembrane transporter activity / ABC-type polyamine transporter activity / regulation of autophagosome size / extracellular exosome biogenesis / negative regulation of lysosomal protein catabolic process / regulation of chaperone-mediated autophagy / P-type ion transporter activity / regulation of autophagy of mitochondrion / regulation of lysosomal protein catabolic process / intracellular monoatomic cation homeostasis / autophagosome-lysosome fusion / autophagosome organization / protein localization to lysosome / positive regulation of exosomal secretion / phosphatidic acid binding / ATPase-coupled monoatomic cation transmembrane transporter activity / multivesicular body membrane / intracellular zinc ion homeostasis / cupric ion binding / regulation of protein localization to nucleus / regulation of mitochondrion organization / phosphatidylinositol-3,5-bisphosphate binding / Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate / lysosomal transport / regulation of intracellular protein transport / lipid homeostasis / cellular response to zinc ion / autophagosome membrane / Ion transport by P-type ATPases / regulation of macroautophagy / transport vesicle / cellular response to manganese ion / multivesicular body / lysosomal lumen / autophagosome / regulation of neuron apoptotic process / positive regulation of protein secretion / autophagy / intracellular calcium ion homeostasis / late endosome / late endosome membrane / manganese ion binding / cellular response to oxidative stress / monoatomic ion transmembrane transport / vesicle / intracellular iron ion homeostasis / lysosome / neuron projection / lysosomal membrane / neuronal cell body / positive regulation of gene expression / ATP hydrolysis activity / zinc ion binding / ATP binding / membrane Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
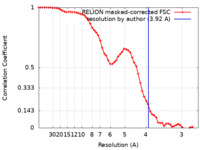
| Method | single particle reconstruction / cryo EM / Resolution: 3.92 Å | ||||||||||||
 Authors Authors | Tomita A / Yamashita K / Nishizawa T / Nureki O | ||||||||||||
| Funding support |  Japan, 3 items Japan, 3 items
| ||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2021 Journal: Mol Cell / Year: 2021Title: Cryo-EM reveals mechanistic insights into lipid-facilitated polyamine export by human ATP13A2. Authors: Atsuhiro Tomita / Takashi Daiho / Tsukasa Kusakizako / Keitaro Yamashita / Satoshi Ogasawara / Takeshi Murata / Tomohiro Nishizawa / Osamu Nureki /   Abstract: The cytoplasmic polyamine maintains cellular homeostasis by chelating toxic metal cations, regulating transcriptional activity, and protecting DNA. ATP13A2 was identified as a lysosomal polyamine ...The cytoplasmic polyamine maintains cellular homeostasis by chelating toxic metal cations, regulating transcriptional activity, and protecting DNA. ATP13A2 was identified as a lysosomal polyamine exporter responsible for polyamine release into the cytosol, and its dysfunction is associated with Alzheimer's disease and other neural degradation diseases. ATP13A2 belongs to the P5 subfamily of the P-type ATPase family, but its mechanisms remain unknown. Here, we report the cryoelectron microscopy (cryo-EM) structures of human ATP13A2 under four different conditions, revealing the structural coupling between the polyamine binding and the dephosphorylation. Polyamine is bound at the luminal tunnel and recognized through numerous electrostatic and π-cation interactions, explaining its broad specificity. The unique N-terminal domain is anchored to the lipid membrane to stabilize the E2P conformation, thereby accelerating the E1P-to-E2P transition. These findings reveal the distinct mechanism of P5B ATPases, thereby paving the way for neuroprotective therapy by activating ATP13A2. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32068.map.gz emd_32068.map.gz | 2.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32068-v30.xml emd-32068-v30.xml emd-32068.xml emd-32068.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32068_fsc.xml emd_32068_fsc.xml | 5.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_32068.png emd_32068.png | 129.1 KB | ||
| Masks |  emd_32068_msk_1.map emd_32068_msk_1.map | 14.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-32068.cif.gz emd-32068.cif.gz | 6.6 KB | ||
| Others |  emd_32068_half_map_1.map.gz emd_32068_half_map_1.map.gz emd_32068_half_map_2.map.gz emd_32068_half_map_2.map.gz | 11.3 MB 11.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32068 http://ftp.pdbj.org/pub/emdb/structures/EMD-32068 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32068 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32068 | HTTPS FTP |
-Related structure data
| Related structure data |  7vpkMC  7vpiC  7vpjC  7vplC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10975 (Title: Cryo-EM structure of the human ATP13A2 / Data size: 3.8 TB EMPIAR-10975 (Title: Cryo-EM structure of the human ATP13A2 / Data size: 3.8 TBData #1: Unaligned movies for E1-ATP state [micrographs - multiframe] Data #2: Unaligned movies for E1P-ADP state [micrographs - multiframe] Data #3: Unaligned movies for SPM-bound E2P state [micrographs - multiframe] Data #4: Unaligned movies for SPM-bound E2Pi state [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32068.map.gz / Format: CCP4 / Size: 14.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32068.map.gz / Format: CCP4 / Size: 14.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | postprocess_masked.mrc from Relion3.1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34875 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_32068_msk_1.map emd_32068_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: run half1 class001.mrc from Relion3.1
| File | emd_32068_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | run_half1_class001.mrc from Relion3.1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: run half1 class002.mrc from Relion3.1
| File | emd_32068_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | run_half1_class002.mrc from Relion3.1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ATP13A2
| Entire | Name: ATP13A2 |
|---|---|
| Components |
|
-Supramolecule #1: ATP13A2
| Supramolecule | Name: ATP13A2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: Human ATP13A2 in complex with SPM and BeF |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Polyamine-transporting ATPase 13A2
| Macromolecule | Name: Polyamine-transporting ATPase 13A2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 129.313391 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GPSRMSADSS PLVGSTPTGY GTLTIGTSID PLSSSVSSVR LSGYCGSPWR VIGYHVVVWM MAGIPLLLFR WKPLWGVRLR LRPCNLAHA ETLVIEIRDK EDSSWQLFTV QVQTEAIGEG SLEPSPQSQA EDGRSQAAVG AVPEGAWKDT AQLHKSEEAV S VGQKRVLR ...String: GPSRMSADSS PLVGSTPTGY GTLTIGTSID PLSSSVSSVR LSGYCGSPWR VIGYHVVVWM MAGIPLLLFR WKPLWGVRLR LRPCNLAHA ETLVIEIRDK EDSSWQLFTV QVQTEAIGEG SLEPSPQSQA EDGRSQAAVG AVPEGAWKDT AQLHKSEEAV S VGQKRVLR YYLFQGQRYI WIETQQAFYQ VSLLDHGRSC DDVHRSRHGL SLQDQMVRKA IYGPNVISIP VKSYPQLLVD EA LNPYYGF QAFSIALWLA DHYYWYALCI FLISSISICL SLYKTRKQSQ TLRDMVKLSM RVCVCRPGGE EEWVDSSELV PGD CLVLPQ EGGLMPCDAA LVAGECMVNE SSLTGESIPV LKTALPEGLG PYCAETHRRH TLFCGTLILQ ARAYVGPHVL AVVT RTGFC TAKGGLVSSI LHPRPINFKF YKHSMKFVAA LSVLALLGTI YSIFILYRNR VPLNEIVIRA LDLVTVVVPP ALPAA MTVC TLYAQSRLRR QGIFCIHPLR INLGGKLQLV CFDKTGTLTE DGLDVMGVVP LKGQAFLPLV PEPRRLPVGP LLRALA TCH ALSRLQDTPV GDPMDLKMVE STGWVLEEEP AADSAFGTQV LAVMRPPLWE PQLQAMEEPP VPVSVLHRFP FSSALQR MS VVVAWPGATQ PEAYVKGSPE LVAGLCNPET VPTDFAQMLQ SYTAAGYRVV ALASKPLPTV PSLEAAQQLT RDTVEGDL S LLGLLVMRNL LKPQTTPVIQ ALRRTRIRAV MVTGDNLQTA VTVARGCGMV APQEHLIIVH ATHPERGQPA SLEFLPMES PTAVNGVKDP DQAASYTVEP DPRSRHLALS GPTFGIIVKH FPKLLPKVLV QGTVFARMAP EQKTELVCEL QKLQYCVGMC GDGANDCGA LKAADVGISL SQAEASVVSP FTSSMASIEC VPMVIREGRC SLDTSFSVFK YMALYSLTQF ISVLILYTIN T NLGDLQFL AIDLVITTTV AVLMSRTGPA LVLGRVRPPG ALLSVPVLSS LLLQMVLVTG VQLGGYFLTL AQPWFVPLNR TV AAPDNLP NYENTVVFSL SSFQYLILAA AVSKGAPFRR PLYTNVPFLV ALALLSSVLV GLVLVPGLLQ GPLALRNITD TGF KLLLLG LVTLNFVGAF MLESVLDQCL PACLRRLRPK RASKKRFKQL ERELAEQPWP PLPAGPLR UniProtKB: Polyamine-transporting ATPase 13A2 |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: BERYLLIUM TRIFLUORIDE ION
| Macromolecule | Name: BERYLLIUM TRIFLUORIDE ION / type: ligand / ID: 4 / Number of copies: 1 / Formula: BEF |
|---|---|
| Molecular weight | Theoretical: 66.007 Da |
| Chemical component information |  ChemComp-BEF: |
-Macromolecule #5: SPERMINE
| Macromolecule | Name: SPERMINE / type: ligand / ID: 5 / Number of copies: 1 / Formula: SPM |
|---|---|
| Molecular weight | Theoretical: 202.34 Da |
| Chemical component information |  ChemComp-SPM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER/RHODIUM / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7vpk: |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)