+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3161 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Campylobacter jejuni pflB deletion bacterial flagellar motor | |||||||||
 Map data Map data | Campylobacter jejuni pflB deletion bacterial flagellar motor | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | bacterial flagellar motor / flagellar diversity / electron cryo-tomography / Campylobacter jejuni / Vibrio fischeri / Salmonella enterica | |||||||||
| Biological species |  | |||||||||
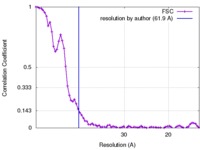
| Method | subtomogram averaging / cryo EM / Resolution: 61.9 Å | |||||||||
 Authors Authors | Beeby M | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2016 Journal: Proc Natl Acad Sci U S A / Year: 2016Title: Diverse high-torque bacterial flagellar motors assemble wider stator rings using a conserved protein scaffold. Authors: Morgan Beeby / Deborah A Ribardo / Caitlin A Brennan / Edward G Ruby / Grant J Jensen / David R Hendrixson /   Abstract: Although it is known that diverse bacterial flagellar motors produce different torques, the mechanism underlying torque variation is unknown. To understand this difference better, we combined genetic ...Although it is known that diverse bacterial flagellar motors produce different torques, the mechanism underlying torque variation is unknown. To understand this difference better, we combined genetic analyses with electron cryo-tomography subtomogram averaging to determine in situ structures of flagellar motors that produce different torques, from Campylobacter and Vibrio species. For the first time, to our knowledge, our results unambiguously locate the torque-generating stator complexes and show that diverse high-torque motors use variants of an ancestrally related family of structures to scaffold incorporation of additional stator complexes at wider radii from the axial driveshaft than in the model enteric motor. We identify the protein components of these additional scaffold structures and elucidate their sequential assembly, demonstrating that they are required for stator-complex incorporation. These proteins are widespread, suggesting that different bacteria have tailored torques to specific environments by scaffolding alternative stator placement and number. Our results quantitatively account for different motor torques, complete the assignment of the locations of the major flagellar components, and provide crucial constraints for understanding mechanisms of torque generation and the evolution of multiprotein complexes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3161.map.gz emd_3161.map.gz | 22.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3161-v30.xml emd-3161-v30.xml emd-3161.xml emd-3161.xml | 7.9 KB 7.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3161_fsc.xml emd_3161_fsc.xml | 7.9 KB | Display |  FSC data file FSC data file |
| Images |  EMD-3161_Campy_pflB.png EMD-3161_Campy_pflB.png | 89.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3161 http://ftp.pdbj.org/pub/emdb/structures/EMD-3161 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3161 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3161 | HTTPS FTP |
-Validation report
| Summary document |  emd_3161_validation.pdf.gz emd_3161_validation.pdf.gz | 254.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3161_full_validation.pdf.gz emd_3161_full_validation.pdf.gz | 253.5 KB | Display | |
| Data in XML |  emd_3161_validation.xml.gz emd_3161_validation.xml.gz | 9.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3161 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3161 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3161 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3161 | HTTPS FTP |
-Related structure data
| Related structure data |  3150C  3154C  3155C  3156C  3157C  3158C  3159C  3160C  3162C  3813C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3161.map.gz / Format: CCP4 / Size: 24 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3161.map.gz / Format: CCP4 / Size: 24 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Campylobacter jejuni pflB deletion bacterial flagellar motor | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 8.122 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Campylobacter jejuni pflB deletion bacterial flagellar motor
| Entire | Name: Campylobacter jejuni pflB deletion bacterial flagellar motor |
|---|---|
| Components |
|
-Supramolecule #1000: Campylobacter jejuni pflB deletion bacterial flagellar motor
| Supramolecule | Name: Campylobacter jejuni pflB deletion bacterial flagellar motor type: sample / ID: 1000 / Oligomeric state: 1 / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 1 MDa / Theoretical: 1 MDa |
-Supramolecule #1: Bacterial flagellar motor
| Supramolecule | Name: Bacterial flagellar motor / type: organelle_or_cellular_component / ID: 1 / Number of copies: 1 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: ETHANE-PROPANE MIXTURE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK III / Method: Variable |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Date | Jan 1, 2015 |
| Image recording | Category: CCD / Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 120 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: -4.0 µm / Nominal defocus min: -4.0 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Nitrogen cooled Gatan 914 / Specimen holder model: SIDE ENTRY, EUCENTRIC / Tilt series - Axis1 - Min angle: -60 ° / Tilt series - Axis1 - Max angle: 60 ° |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)