[English] 日本語
 Yorodumi
Yorodumi- EMDB-31558: Structure of human SGLT2-MAP17 complex bound with empagliflozin -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31558 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human SGLT2-MAP17 complex bound with empagliflozin | ||||||||||||
 Map data Map data | postprocessed map reboxed to 100 | ||||||||||||
 Sample Sample |
| ||||||||||||
| Function / homology |  Function and homology information Function and homology informationlow-affinity D-glucose:sodium symporter activity / Defective SLC5A2 causes renal glucosuria (GLYS1) / alpha-glucoside transport / D-glucose:sodium symporter activity / alpha-glucoside transmembrane transporter activity / hexose transmembrane transport / D-glucose transmembrane transporter activity / renal D-glucose absorption / Cellular hexose transport / D-glucose import across plasma membrane ...low-affinity D-glucose:sodium symporter activity / Defective SLC5A2 causes renal glucosuria (GLYS1) / alpha-glucoside transport / D-glucose:sodium symporter activity / alpha-glucoside transmembrane transporter activity / hexose transmembrane transport / D-glucose transmembrane transporter activity / renal D-glucose absorption / Cellular hexose transport / D-glucose import across plasma membrane / sodium ion import across plasma membrane / sodium ion transport / carbohydrate metabolic process / apical plasma membrane / extracellular exosome / membrane / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.95 Å | ||||||||||||
 Authors Authors | Chen L / Niu Y / Liu R | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Structural basis of inhibition of the human SGLT2-MAP17 glucose transporter. Authors: Yange Niu / Rui Liu / Chengcheng Guan / Yuan Zhang / Zhixing Chen / Stefan Hoerer / Herbert Nar / Lei Chen /   Abstract: Human sodium-glucose cotransporter 2 (hSGLT2) mediates the reabsorption of the majority of filtrated glucose in the kidney. Pharmacological inhibition of hSGLT2 by oral small-molecule inhibitors, ...Human sodium-glucose cotransporter 2 (hSGLT2) mediates the reabsorption of the majority of filtrated glucose in the kidney. Pharmacological inhibition of hSGLT2 by oral small-molecule inhibitors, such as empagliflozin, leads to enhanced excretion of glucose and is widely used in the clinic to manage blood glucose levels for the treatment of type 2 diabetes. Here we determined the cryogenic electron microscopy structure of the hSGLT2-MAP17 complex in the empagliflozin-bound state to an overall resolution of 2.95 Å. Our structure shows eukaryotic SGLT-specific structural features. MAP17 interacts with transmembrane helix 13 of hSGLT2. Empagliflozin occupies both the sugar-substrate-binding site and the external vestibule to lock hSGLT2 in an outward-open conformation, thus inhibiting the transport cycle. Our work provides a framework for understanding the mechanism of SLC5A family glucose transporters and also develops a foundation for the future rational design and optimization of new inhibitors targeting these transporters. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31558.map.gz emd_31558.map.gz | 3.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31558-v30.xml emd-31558-v30.xml emd-31558.xml emd-31558.xml | 14.4 KB 14.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31558_fsc.xml emd_31558_fsc.xml | 10 KB | Display |  FSC data file FSC data file |
| Images |  emd_31558.png emd_31558.png | 68.5 KB | ||
| Others |  emd_31558_additional_1.map.gz emd_31558_additional_1.map.gz | 65.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31558 http://ftp.pdbj.org/pub/emdb/structures/EMD-31558 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31558 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31558 | HTTPS FTP |
-Validation report
| Summary document |  emd_31558_validation.pdf.gz emd_31558_validation.pdf.gz | 485.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31558_full_validation.pdf.gz emd_31558_full_validation.pdf.gz | 484.9 KB | Display | |
| Data in XML |  emd_31558_validation.xml.gz emd_31558_validation.xml.gz | 10.2 KB | Display | |
| Data in CIF |  emd_31558_validation.cif.gz emd_31558_validation.cif.gz | 13.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31558 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31558 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31558 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31558 | HTTPS FTP |
-Related structure data
| Related structure data |  7vsiMC  7fen M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31558.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31558.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | postprocessed map reboxed to 100 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.821 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
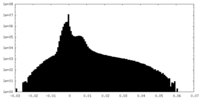
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: refined map from relion
| File | emd_31558_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | refined map from relion | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : human soluble guanylate cyclase
| Entire | Name: human soluble guanylate cyclase |
|---|---|
| Components |
|
-Supramolecule #1: human soluble guanylate cyclase
| Supramolecule | Name: human soluble guanylate cyclase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Sodium/glucose cotransporter 2
| Macromolecule | Name: Sodium/glucose cotransporter 2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 72.949414 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEEHTEAGSA PEMGAQKALI DNPADILVIA AYFLLVIGVG LWSMCRTNRG TVGGYFLAGR SMVWWPVGAS LFASNIGSGH FVGLAGTGA ASGLAVAGFE WNALFVVLLL GWLFAPVYLT AGVITMPQYL RKRFGGRRIR LYLSVLSLFL YIFTKISVDM F SGAVFIQQ ...String: MEEHTEAGSA PEMGAQKALI DNPADILVIA AYFLLVIGVG LWSMCRTNRG TVGGYFLAGR SMVWWPVGAS LFASNIGSGH FVGLAGTGA ASGLAVAGFE WNALFVVLLL GWLFAPVYLT AGVITMPQYL RKRFGGRRIR LYLSVLSLFL YIFTKISVDM F SGAVFIQQ ALGWNIYASV IALLGITMIY TVTGGLAALM YTDTVQTFVI LGGACILMGY AFHEVGGYSG LFDKYLGAAT SL TVSEDPA VGNISSFCYR PRPDSYHLLR HPVTGDLPWP ALLLGLTIVS GWYWCSDQVI VQRCLAGKSL THIKAGCILC GYL KLTPMF LMVMPGMISR ILYPDEVACV VPEVCRRVCG TEVGCSNIAY PRLVVKLMPN GLRGLMLAVM LAALMSSLAS IFNS SSTLF TMDIYTRLRP RAGDRELLLV GRLWVVFIVV VSVAWLPVVQ AAQGGQLFDY IQAVSSYLAP PVSAVFVLAL FVPRV NEQG AFWGLIGGLL MGLARLIPEF SFGSGSCVQP SACPAFLCGV HYLYFAIVLF FCSGLLTLTV SLCTAPIPRK HLHRLV FSL RHSKEEREDL DADEQQGSSL PVQNGCPESA MEMNEPQAPA PSLFRQCLLW FCGMSRGGVG SPPPLTQEEA AAAARRL ED ISEDPSWARV VNLNALLMMA VAVFLWGFYA |
-Macromolecule #2: PDZK1-interacting protein 1
| Macromolecule | Name: PDZK1-interacting protein 1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 5.937134 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSALSLLILG LLTAVPPASC QQGLGNLQPW MQGLIAVAVF LVLVAIAFAV NHFWCQ |
-Macromolecule #3: PALMITIC ACID
| Macromolecule | Name: PALMITIC ACID / type: ligand / ID: 3 / Number of copies: 2 / Formula: PLM |
|---|---|
| Molecular weight | Theoretical: 256.424 Da |
| Chemical component information |  ChemComp-PLM: |
-Macromolecule #4: (2S,3R,4R,5S,6R)-2-[4-chloranyl-3-[[4-[(3S)-oxolan-3-yl]oxyphenyl...
| Macromolecule | Name: (2S,3R,4R,5S,6R)-2-[4-chloranyl-3-[[4-[(3S)-oxolan-3-yl]oxyphenyl]methyl]phenyl]-6-(hydroxymethyl)oxane-3,4,5-triol type: ligand / ID: 4 / Number of copies: 1 / Formula: 7R3 |
|---|---|
| Molecular weight | Theoretical: 450.909 Da |
| Chemical component information | 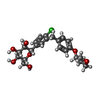 ChemComp-7R3: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)