+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31433 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of human Kv4.2-KChIP1 complex | |||||||||
 Map data Map data | map A | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | complex / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationKv4.3-KChIP1 channel complex / Kv4.2-KChIP2 channel complex / A-type (transient outward) potassium channel activity / Phase 1 - inactivation of fast Na+ channels / voltage-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / potassium ion export across plasma membrane / membrane repolarization / Voltage gated Potassium channels / regulation of potassium ion transmembrane transport / anchoring junction ...Kv4.3-KChIP1 channel complex / Kv4.2-KChIP2 channel complex / A-type (transient outward) potassium channel activity / Phase 1 - inactivation of fast Na+ channels / voltage-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / potassium ion export across plasma membrane / membrane repolarization / Voltage gated Potassium channels / regulation of potassium ion transmembrane transport / anchoring junction / postsynaptic specialization membrane / neuronal cell body membrane / locomotor rhythm / regulation of heart contraction / action potential / voltage-gated potassium channel activity / plasma membrane raft / regulation of signal transduction / potassium channel activity / potassium channel regulator activity / neuronal action potential / voltage-gated potassium channel complex / potassium ion transmembrane transport / sensory perception of pain / muscle contraction / protein homooligomerization / GABA-ergic synapse / cytoplasmic side of plasma membrane / cellular response to hypoxia / dendritic spine / chemical synaptic transmission / perikaryon / transmembrane transporter binding / postsynaptic membrane / neuronal cell body / calcium ion binding / synapse / dendrite / glutamatergic synapse / metal ion binding / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
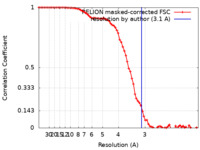
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Kise Y / Nureki O | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Structural basis of gating modulation of Kv4 channel complexes. Authors: Yoshiaki Kise / Go Kasuya / Hiroyuki H Okamoto / Daichi Yamanouchi / Kan Kobayashi / Tsukasa Kusakizako / Tomohiro Nishizawa / Koichi Nakajo / Osamu Nureki /  Abstract: Modulation of voltage-gated potassium (Kv) channels by auxiliary subunits is central to the physiological function of channels in the brain and heart. Native Kv4 tetrameric channels form ...Modulation of voltage-gated potassium (Kv) channels by auxiliary subunits is central to the physiological function of channels in the brain and heart. Native Kv4 tetrameric channels form macromolecular ternary complexes with two auxiliary β-subunits-intracellular Kv channel-interacting proteins (KChIPs) and transmembrane dipeptidyl peptidase-related proteins (DPPs)-to evoke rapidly activating and inactivating A-type currents, which prevent the backpropagation of action potentials. However, the modulatory mechanisms of Kv4 channel complexes remain largely unknown. Here we report cryo-electron microscopy structures of the Kv4.2-DPP6S-KChIP1 dodecamer complex, the Kv4.2-KChIP1 and Kv4.2-DPP6S octamer complexes, and Kv4.2 alone. The structure of the Kv4.2-KChIP1 complex reveals that the intracellular N terminus of Kv4.2 interacts with its C terminus that extends from the S6 gating helix of the neighbouring Kv4.2 subunit. KChIP1 captures both the N and the C terminus of Kv4.2. In consequence, KChIP1 would prevent N-type inactivation and stabilize the S6 conformation to modulate gating of the S6 helices within the tetramer. By contrast, unlike the reported auxiliary subunits of voltage-gated channel complexes, DPP6S interacts with the S1 and S2 helices of the Kv4.2 voltage-sensing domain, which suggests that DPP6S stabilizes the conformation of the S1-S2 helices. DPP6S may therefore accelerate the voltage-dependent movement of the S4 helices. KChIP1 and DPP6S do not directly interact with each other in the Kv4.2-KChIP1-DPP6S ternary complex. Thus, our data suggest that two distinct modes of modulation contribute in an additive manner to evoke A-type currents from the native Kv4 macromolecular complex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31433.map.gz emd_31433.map.gz | 7.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31433-v30.xml emd-31433-v30.xml emd-31433.xml emd-31433.xml | 18.4 KB 18.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31433_fsc.xml emd_31433_fsc.xml | 7.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_31433.png emd_31433.png | 64.3 KB | ||
| Masks |  emd_31433_msk_1.map emd_31433_msk_1.map | 36.3 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-31433.cif.gz emd-31433.cif.gz | 6.2 KB | ||
| Others |  emd_31433_half_map_1.map.gz emd_31433_half_map_1.map.gz emd_31433_half_map_2.map.gz emd_31433_half_map_2.map.gz | 24.7 MB 24.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31433 http://ftp.pdbj.org/pub/emdb/structures/EMD-31433 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31433 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31433 | HTTPS FTP |
-Related structure data
| Related structure data |  7f3fMC  7e7zC  7e83C  7e84C  7e87C  7e89C  7e8bC  7e8eC  7e8gC  7e8hC  7f0jC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31433.map.gz / Format: CCP4 / Size: 36.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31433.map.gz / Format: CCP4 / Size: 36.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map A | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.00226 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_31433_msk_1.map emd_31433_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_31433_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_31433_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : human Kv4.2-KChIP1 complex
| Entire | Name: human Kv4.2-KChIP1 complex |
|---|---|
| Components |
|
-Supramolecule #1: human Kv4.2-KChIP1 complex
| Supramolecule | Name: human Kv4.2-KChIP1 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #2: human Kv4.2-KChIP1 complex
| Supramolecule | Name: human Kv4.2-KChIP1 complex / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Potassium voltage-gated channel subfamily D member 2
| Macromolecule | Name: Potassium voltage-gated channel subfamily D member 2 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 70.57543 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAAGVAAWLP FARAAAIGWM PVASGPMPAP PRQERKRTQD ALIVLNVSGT RFQTWQDTLE RYPDTLLGSS ERDFFYHPET QQYFFDRDP DIFRHILNFY RTGKLHYPRH ECISAYDEEL AFFGLIPEII GDCCYEEYKD RRRENAERLQ DDADTDTAGE S ALPTMTAR ...String: MAAGVAAWLP FARAAAIGWM PVASGPMPAP PRQERKRTQD ALIVLNVSGT RFQTWQDTLE RYPDTLLGSS ERDFFYHPET QQYFFDRDP DIFRHILNFY RTGKLHYPRH ECISAYDEEL AFFGLIPEII GDCCYEEYKD RRRENAERLQ DDADTDTAGE S ALPTMTAR QRVWRAFENP HTSTMALVFY YVTGFFIAVS VIANVVETVP CGSSPGHIKE LPCGERYAVA FFCLDTACVM IF TVEYLLR LAAAPSRYRF VRSVMSIIDV VAILPYYIGL VMTDNEDVSG AFVTLRVFRV FRIFKFSRHS QGLRILGYTL KSC ASELGF LLFSLTMAII IFATVMFYAE KGSSASKFTS IPAAFWYTIV TMTTLGYGDM VPKTIAGKIF GSICSLSGVL VIAL PVPVI VSNFSRIYHQ NQRADKRRAQ KKARLARIRA AKSGSANAYM QSKRSGLLSN QLQSSEDEPA FVSKSGSSFE TQHHH LLHC LEKTTNHEFV DEQVFEESCM EVATVNRPSS HSPSLSSQQG VTSTCCSRRH KKTFRIPNAN VSGSHRGSVQ ELSTIQ IRC VERTPLSNSR SSLNAKMEEC VKLNCEQPYV TTAIISIPTP PVTTPEGDDR PESPEYSGGN IVRVSAL UniProtKB: A-type voltage-gated potassium channel KCND2 |
-Macromolecule #2: Isoform 2 of Kv channel-interacting protein 1
| Macromolecule | Name: Isoform 2 of Kv channel-interacting protein 1 / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.27367 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGAVMGTFSS LQTKQRRPSK DKIEDELEMT MVCHRPEGLE QLEAQTNFTK RELQVLYRGF KNECPSGVVN EDTFKQIYAQ FFPHGDAST YAHYLFNAFD TTQTGSVKFE DFVTALSILL RGTVHEKLRW TFNLYDINKD GYINKEEMMD IVKAIYDMMG K YTYPVLKE ...String: MGAVMGTFSS LQTKQRRPSK DKIEDELEMT MVCHRPEGLE QLEAQTNFTK RELQVLYRGF KNECPSGVVN EDTFKQIYAQ FFPHGDAST YAHYLFNAFD TTQTGSVKFE DFVTALSILL RGTVHEKLRW TFNLYDINKD GYINKEEMMD IVKAIYDMMG K YTYPVLKE DTPRQHVDVF FQKMDKNKDG IVTLDEFLES CQEDDNIMRS LQLFQNVM UniProtKB: A-type potassium channel modulatory protein KCNIP1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)