[日本語] English
 万見
万見- EMDB-31100: SARS-CoV2 Spike Protein structure at pH 7.4 with C1 Symmetry (Class 9) -
+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-31100 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | SARS-CoV2 Spike Protein structure at pH 7.4 with C1 Symmetry (Class 9) | ||||||||||||||||||||||||
 マップデータ マップデータ | SARS-CoV2 Spike Protein structure at pH 7.4 with C1 Symmetry (Class 9) | ||||||||||||||||||||||||
 試料 試料 |
| ||||||||||||||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell / membrane fusion / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / receptor ligand activity / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane 類似検索 - 分子機能 | ||||||||||||||||||||||||
| 生物種 |  | ||||||||||||||||||||||||
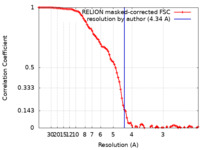
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 4.34 Å | ||||||||||||||||||||||||
 データ登録者 データ登録者 | Pramanick I / Sengupta N / Mishra S / Pandey S / Girish N / Das A / Dutta S | ||||||||||||||||||||||||
| 資金援助 |  インド, 7件 インド, 7件
| ||||||||||||||||||||||||
 引用 引用 |  ジャーナル: Structure / 年: 2021 ジャーナル: Structure / 年: 2021タイトル: Conformational flexibility and structural variability of SARS-CoV2 S protein. 著者: Ishika Pramanick / Nayanika Sengupta / Suman Mishra / Suman Pandey / Nidhi Girish / Alakta Das / Somnath Dutta /  要旨: Spike (S) glycoprotein of SARS-CoV2 exists chiefly in two conformations, open and closed. Most previous structural studies on S protein have been conducted at pH 8.0, but knowledge of the ...Spike (S) glycoprotein of SARS-CoV2 exists chiefly in two conformations, open and closed. Most previous structural studies on S protein have been conducted at pH 8.0, but knowledge of the conformational propensities under both physiological and endosomal pH conditions is important to inform vaccine development. Our current study employed single-particle cryoelectron microscopy to visualize multiple states of open and closed conformations of S protein at physiological pH 7.4 and near-physiological pH 6.5 and pH 8.0. Propensities of open and closed conformations were found to differ with pH changes, whereby around 68% of S protein exists in open conformation at pH 7.4. Furthermore, we noticed a continuous movement in the N-terminal domain, receptor-binding domain (RBD), S2 domain, and stalk domain of S protein conformations at various pH values. Several key residues involving RBD-neutralizing epitopes are differentially exposed in each conformation. This study will assist in developing novel therapeutic measures against SARS-CoV2. | ||||||||||||||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_31100.map.gz emd_31100.map.gz | 59.9 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-31100-v30.xml emd-31100-v30.xml emd-31100.xml emd-31100.xml | 11.4 KB 11.4 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_31100_fsc.xml emd_31100_fsc.xml | 9.2 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_31100.png emd_31100.png | 150.2 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31100 http://ftp.pdbj.org/pub/emdb/structures/EMD-31100 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31100 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31100 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_31100_validation.pdf.gz emd_31100_validation.pdf.gz | 392.6 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_31100_full_validation.pdf.gz emd_31100_full_validation.pdf.gz | 392.1 KB | 表示 | |
| XML形式データ |  emd_31100_validation.xml.gz emd_31100_validation.xml.gz | 11 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31100 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31100 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31100 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31100 | HTTPS FTP |
-関連構造データ
| 関連構造データ | C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_31100.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_31100.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | SARS-CoV2 Spike Protein structure at pH 7.4 with C1 Symmetry (Class 9) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.17 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : SARS-CoV2 Spike Protein structure at pH 7.4 with C1 Symmetry (Class 9)
| 全体 | 名称: SARS-CoV2 Spike Protein structure at pH 7.4 with C1 Symmetry (Class 9) |
|---|---|
| 要素 |
|
-超分子 #1: SARS-CoV2 Spike Protein structure at pH 7.4 with C1 Symmetry (Class 9)
| 超分子 | 名称: SARS-CoV2 Spike Protein structure at pH 7.4 with C1 Symmetry (Class 9) タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: all |
|---|---|
| 由来(天然) | 生物種:  |
| 組換発現 | 生物種:  Homo sapiens (ヒト) / 組換細胞: Mammalian expression Expi293F cells Homo sapiens (ヒト) / 組換細胞: Mammalian expression Expi293F cells |
-分子 #1: SARS-CoV2 Spike Protein structure at pH 7.4 with C1 Symmetry (Class 9)
| 分子 | 名称: SARS-CoV2 Spike Protein structure at pH 7.4 with C1 Symmetry (Class 9) タイプ: other / ID: 1 / 分類: other |
|---|---|
| 由来(天然) | 生物種:  |
| 配列 | 文字列: MGILPSPGMP ALLSLVSLLS VLLMGCVAET GTQCVNLTTR TQLPPAYTNS FTRGVYYPDK VFRSSVLHST QDLFLPFFSN VTWFHAIHVS GTNGTKRFDN PVLPFNDGVY FASTEKSNII RGWIFGTTLD SKTQSLLIVN NATNVVIKVC EFQFCNDPFL GVYYHKNNKS ...文字列: MGILPSPGMP ALLSLVSLLS VLLMGCVAET GTQCVNLTTR TQLPPAYTNS FTRGVYYPDK VFRSSVLHST QDLFLPFFSN VTWFHAIHVS GTNGTKRFDN PVLPFNDGVY FASTEKSNII RGWIFGTTLD SKTQSLLIVN NATNVVIKVC EFQFCNDPFL GVYYHKNNKS WMESEFRVYS SANNCTFEYV SQPFLMDLEG KQGNFKNLRE FVFKNIDGYF KIYSKHTPIN LVRDLPQGFS ALEPLVDLPI GINITRFQTL LALHRSYLTP GDSSSGWTAG AAAYYVGYLQ PRTFLLKYNE NGTITDAVDC ALDPLSETKC TLKSFTVEKG IYQTSNFRVQ PTESIVRFPN ITNLCPFGEV FNATRFASVY AWNRKRISNC VADYSVLYNS ASFSTFKCYG VSPTKLNDLC FTNVYADSFV IRGDEVRQIA PGQTGKIADY NYKLPDDFTG CVIAWNSNNL DSKVGGNYNY LYRLFRKSNL KPFERDISTE IYQAGSTPCN GVEGFNCYFP LQSYGFQPTN GVGYQPYRVV VLSFELLHAP ATVCGPKKST NLVKNKCVNF NFNGLTGTGV LTESNKKFLP FQQFGRDIAD TTDAVRDPQT LEILDITPCS FGGVSVITPG TNTSNQVAVL YQDVNCTEVP VAIHADQLTP TWRVYSTGSN VFQTRAGCLI GAEHVNNSYE CDIPIGAGIC ASYQTQTNSP SGAGSVASQS IIAYTMSLGA ENSVAYSNNS IAIPTNFTIS VTTEILPVSM TKTSVDCTMY ICGDSTECSN LLLQYGSFCT QLNRALTGIA VEQDKNTQEV FAQVKQIYKT PPIKDFGGFN FSQILPDPSK PSKRSFIEDL LFNKVTLADA GFIKQYGDCL GDIAARDLIC AQKFNGLTVL PPLLTDEMIA QYTSALLAGT ITSGWTFGAG AALQIPFAMQ MAYRFNGIGV TQNVLYENQK LIANQFNSAI GKIQDSLSST ASALGKLQDV VNQNAQALNT LVKQLSSNFG AISSVLNDIL SRLDPPEAEV QIDRLITGRL QSLQTYVTQQ LIRAAEIRAS ANLAATKMSE CVLGQSKRVD FCGKGYHLMS FPQSAPHGVV FLHVTYVPAQ EKNFTTAPAI CHDGKAHFPR EGVFVSNGTH WFVTQRNFYE PQIITTDNTF VSGNCDVVIG IVNNTVYDPL QPELDSFKEE LDKYFKNHTS PDVDLGDISG INASVVNIQK EIDRLNEVAK NLNESLIDLQ ELGKYEQYIK GSGRENLYFQ GGGGSGYIPE APRDGQAYVR KDGEWVLLST FLGHHHHHHH H |
| 組換発現 | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.4 |
|---|---|
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TALOS ARCTICA |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 検出モード: COUNTING / 平均電子線量: 80.0 e/Å2 |
| 電子線 | 加速電圧: 200 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: OTHER / 撮影モード: BRIGHT FIELD |
| 実験機器 |  モデル: Talos Arctica / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)