[English] 日本語
 Yorodumi
Yorodumi- EMDB-3074: Novel inter-subunit contacts in Barley Stripe Mosaic Virus reveal... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3074 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Novel inter-subunit contacts in Barley Stripe Mosaic Virus revealed by cryo-EM | |||||||||
 Map data Map data | Reconstruction of the narrow Capsid of BSMV | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | BSMV / virus / helical / cryo-EM / image processing / MSA | |||||||||
| Function / homology | Tobacco mosaic virus-like, coat protein / Tobacco mosaic virus-like, coat protein superfamily / Virus coat protein (TMV like) / helical viral capsid / structural molecule activity / identical protein binding / Capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |   Tobacco mosaic virus / Tobacco mosaic virus /  Barley stripe mosaic virus Barley stripe mosaic virus | |||||||||
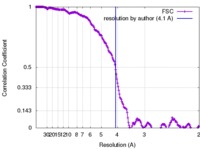
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | Clare DK / Pechnikova E / Skurat E / Makarov V / Sokolova OS / Solovyev AG / Orlova EV | |||||||||
 Citation Citation |  Journal: Structure / Year: 2015 Journal: Structure / Year: 2015Title: Novel Inter-Subunit Contacts in Barley Stripe Mosaic Virus Revealed by Cryo-Electron Microscopy. Authors: Daniel Kofi Clare / Eugenia V Pechnikova / Eugene V Skurat / Valentin V Makarov / Olga S Sokolova / Andrey G Solovyev / Elena V Orlova /   Abstract: Barley stripe mosaic virus (BSMV, genus Hordeivirus) is a rod-shaped single-stranded RNA virus similar to viruses of the structurally characterized and well-studied genus Tobamovirus. Here we report ...Barley stripe mosaic virus (BSMV, genus Hordeivirus) is a rod-shaped single-stranded RNA virus similar to viruses of the structurally characterized and well-studied genus Tobamovirus. Here we report the first high-resolution structure of BSMV at 4.1 Å obtained by cryo-electron microscopy. We discovered that BSMV forms two types of virion that differ in the number of coat protein (CP) subunits per turn and interactions between the CP subunits. While BSMV and tobacco mosaic virus CP subunits have a similar fold and interact with RNA using conserved residues, the axial contacts between the CP of these two viral groups are considerably different. BSMV CP subunits lack substantial axial contacts and are held together by a previously unobserved lateral contact formed at the virion surface via an interacting loop, which protrudes from the CP hydrophobic core to the adjacent CP subunit. These data provide an insight into diversity in structural organization of helical viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3074.map.gz emd_3074.map.gz | 97.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3074-v30.xml emd-3074-v30.xml emd-3074.xml emd-3074.xml | 12.6 KB 12.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3074_fsc.xml emd_3074_fsc.xml | 20 KB | Display |  FSC data file FSC data file |
| Images |  emd_3074.jpg emd_3074.jpg | 99.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3074 http://ftp.pdbj.org/pub/emdb/structures/EMD-3074 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3074 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3074 | HTTPS FTP |
-Related structure data
| Related structure data |  5a7aMC  3073C  3077C  3078C  5a79C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3074.map.gz / Format: CCP4 / Size: 100.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3074.map.gz / Format: CCP4 / Size: 100.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of the narrow Capsid of BSMV | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
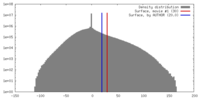
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : chimeric Barley Stripe Mosaic Virus narrow virion
| Entire | Name: chimeric Barley Stripe Mosaic Virus narrow virion |
|---|---|
| Components |
|
-Supramolecule #1000: chimeric Barley Stripe Mosaic Virus narrow virion
| Supramolecule | Name: chimeric Barley Stripe Mosaic Virus narrow virion / type: sample / ID: 1000 Details: The sample is a helical virus with 106 copies of the capsid protein per helical repeat Oligomeric state: Helical, with 106 copies of the capsid protein per helical repeat Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 22.5 KDa / Theoretical: 22.5 KDa |
-Supramolecule #1: Barley stripe mosaic virus
| Supramolecule | Name: Barley stripe mosaic virus / type: virus / ID: 1 / Name.synonym: BSMV / Details: Has RNA bound / NCBI-ID: 12327 / Sci species name: Barley stripe mosaic virus / Sci species strain: ND18 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No / Syn species name: BSMV |
|---|---|
| Host (natural) | Organism:  |
| Host system | Organism:  |
| Molecular weight | Experimental: 22.5 KDa / Theoretical: 22.5 KDa |
| Virus shell | Shell ID: 1 / Name: narrow virion / Diameter: 216 Å |
-Macromolecule #1: RNA
| Macromolecule | Name: RNA / type: rna / ID: 1 / Name.synonym: RNA / Classification: OTHER / Structure: SINGLE STRANDED / Synthetic?: No |
|---|---|
| Source (natural) | Organism:   Tobacco mosaic virus Tobacco mosaic virus |
| Sequence | String: GAA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 50mM Tris-HCL, 50mM KCl, 10mM MgCl2 |
| Grid | Details: c-flats r2/2 or home made continuous carbon grids |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 40 % / Chamber temperature: 100 K / Instrument: HOMEMADE PLUNGER Method: 3.5ul of sample was added to continuous carbon coated or c-flat grids, blotted for 2 seconds and then plunged in liquid ethane |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Min: 80 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 150,000 times magnification |
| Date | Jun 1, 2012 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Digitization - Sampling interval: 15 µm / Number real images: 297 / Average electron dose: 25 e/Å2 / Details: each image was a 1 second low dose exposure / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 150000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.3 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.7 µm / Nominal magnification: 150000 |
| Sample stage | Specimen holder model: GATAN HELIUM |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)