+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30644 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Calcium-Sensing Receptor in complex with NPS-2143 | |||||||||
 Map data Map data | map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Calcium-Sensing Receptor / CaSR / NPS-2143 / MEMBRANE PROTEIN | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Wen TL / Yang X | |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2021 Journal: Sci Adv / Year: 2021Title: Structural basis for activation and allosteric modulation of full-length calcium-sensing receptor. Authors: Tianlei Wen / Ziyu Wang / Xiaozhe Chen / Yue Ren / Xuhang Lu / Yangfei Xing / Jing Lu / Shenghai Chang / Xing Zhang / Yuequan Shen / Xue Yang /  Abstract: Calcium-sensing receptor (CaSR) is a class C G protein-coupled receptor (GPCR) that plays an important role in calcium homeostasis and parathyroid hormone secretion. Here, we present multiple cryo- ...Calcium-sensing receptor (CaSR) is a class C G protein-coupled receptor (GPCR) that plays an important role in calcium homeostasis and parathyroid hormone secretion. Here, we present multiple cryo-electron microscopy structures of full-length CaSR in distinct ligand-bound states. Ligands (Ca and l-tryptophan) bind to the extracellular domain of CaSR and induce large-scale conformational changes, leading to the closure of two heptahelical transmembrane domains (7TMDs) for activation. The positive modulator (evocalcet) and the negative allosteric modulator (NPS-2143) occupy the similar binding pocket in 7TMD. The binding of NPS-2143 causes a considerable rearrangement of two 7TMDs, forming an inactivated TM6/TM6 interface. Moreover, a total of 305 disease-causing missense mutations of CaSR have been mapped to the structure in the active state, creating hotspot maps of five clinical endocrine disorders. Our results provide a structural framework for understanding the activation, allosteric modulation mechanism, and disease therapy for class C GPCRs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30644.map.gz emd_30644.map.gz | 79 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30644-v30.xml emd-30644-v30.xml emd-30644.xml emd-30644.xml | 16 KB 16 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30644.png emd_30644.png | 61.4 KB | ||
| Filedesc metadata |  emd-30644.cif.gz emd-30644.cif.gz | 6 KB | ||
| Others |  emd_30644_additional_1.map.gz emd_30644_additional_1.map.gz emd_30644_additional_2.map.gz emd_30644_additional_2.map.gz | 2.4 MB 64.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30644 http://ftp.pdbj.org/pub/emdb/structures/EMD-30644 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30644 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30644 | HTTPS FTP |
-Related structure data
| Related structure data |  7dd5MC  7dd6C  7dd7C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_30644.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30644.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.014 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
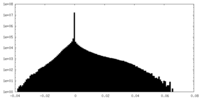
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: TM
| File | emd_30644_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TM | ||||||||||||
| Projections & Slices |
| ||||||||||||
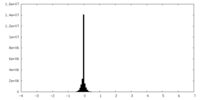
| Density Histograms |
-Additional map: ECD
| File | emd_30644_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ECD | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Calcium-Sensing Receptor from Gallus gallus with Ca2+/L-Trp/NPS-2143
| Entire | Name: Calcium-Sensing Receptor from Gallus gallus with Ca2+/L-Trp/NPS-2143 |
|---|---|
| Components |
|
-Supramolecule #1: Calcium-Sensing Receptor from Gallus gallus with Ca2+/L-Trp/NPS-2143
| Supramolecule | Name: Calcium-Sensing Receptor from Gallus gallus with Ca2+/L-Trp/NPS-2143 type: cell / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Calcium-Sensing Receptor
| Macromolecule | Name: Calcium-Sensing Receptor / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 119.314234 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MTLYSCCLIL LLFTWNTAAY GPNQRAQKKG DIILGGLFPI HFGVAAKDQD LKSRPESVEC IRYNFRGFRW LQAMIFAIEE INNSPNLLP NMTLGYRIFD TCNTVSKALE ATLSFVAQNK IDSLNLDEFC NCSEHIPSTI AVVGATGSGV STAVANLLGL F YIPQVSYA ...String: MTLYSCCLIL LLFTWNTAAY GPNQRAQKKG DIILGGLFPI HFGVAAKDQD LKSRPESVEC IRYNFRGFRW LQAMIFAIEE INNSPNLLP NMTLGYRIFD TCNTVSKALE ATLSFVAQNK IDSLNLDEFC NCSEHIPSTI AVVGATGSGV STAVANLLGL F YIPQVSYA SSSRLLSNKN QFKSFLRTIP NDEHQATAMA DIIEYFRWNW VGTIAADDDY GRPGIEKFRE EAEERDICID FS ELISQYS DEEEIQQVVE VIQNSTARVI VVFSSGPDLE PLIKEIVRRN ITGKIWLASE AWASSSLIAM PEFFRVIGST IGF ALKAGQ IPGFREFLQK VHPKKSANNG FAKEFWEETF NCYLPSESKN SPASASFHKA HEEGLGAGNG TAAFRPPCTG DENI TSVET PYMDFTHLRI SYNVYLAVYS IAHALQDIYT CTPGKGLFTN GSCADIKKVE AWQVLKHLRH LNFTSNMGEQ VDFDE FGDL VGNYSIINWH LSPEDGSVVF EEVGHYNVYA KKGERLFINE NKILWSGFSK EVPFSNCSRD CLPGTRKGII EGEPTC CFE CVDCPDGEYS DETDASACDK CPEDYWSNEN HTSCIPKQIE FLSWTEPFGI ALTLFAVLGI FLTSFVLGVF TKFRNTP IV KATNRELSYL LLFSLLCCFS SSLFFIGEPQ NWTCRLRQPA FGISFVLCIS CILVKTNRVL LVFEAKIPTS LHRKWWGL N LQFLLVFLCT FVQIVICVIW LYTAPPSSYR NHELEDEIIF ITCHEGSLMA LGFLIGYTCL LAAICFFFAF KSRKLPENF NEAKFITFSM LIFFIVWISF IPAYASTYGK FVSAVEVIAI LAASFGLLAC IFFNKVYIIL FKPSRNTIEE VRCSTAAHAF KVAARATLR RSNVSRKRSN SLGGSTGSTP SSSISSKSNH EDPFPLPASA ERQRQQQRGC KQKVSFGSGT VTLSLSFEEP Q KNAMANRN AKRRNSLEAQ NSDDSLMRHR ALLALQNSES LSAEPGFQTA SSPETSSQES VVGDNKEEVP NPEAEPSLPS AN SRNFIGT GGSSVTENTV HSLESALEVL FQ |
-Macromolecule #3: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Macromolecule #4: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 4 / Number of copies: 6 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #6: TRYPTOPHAN
| Macromolecule | Name: TRYPTOPHAN / type: ligand / ID: 6 / Number of copies: 2 / Formula: TRP |
|---|---|
| Molecular weight | Theoretical: 204.225 Da |
| Chemical component information |  ChemComp-TRP: |
-Macromolecule #7: 2-chloro-6-[(2R)-2-hydroxy-3-{[2-methyl-1-(naphthalen-2-yl)propan...
| Macromolecule | Name: 2-chloro-6-[(2R)-2-hydroxy-3-{[2-methyl-1-(naphthalen-2-yl)propan-2-yl]amino}propoxy]benzonitrile type: ligand / ID: 7 / Number of copies: 2 / Formula: YP1 |
|---|---|
| Molecular weight | Theoretical: 408.921 Da |
| Chemical component information |  ChemComp-YP1: |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 2 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.2 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 86921 |
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7dd5: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)