[English] 日本語
 Yorodumi
Yorodumi- EMDB-30572: Structure of COVID-19 RNA-dependent RNA polymerase bound to suramin -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30572 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of COVID-19 RNA-dependent RNA polymerase bound to suramin | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | COVID-19 / RNA polymerase / suramin binding / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein guanylyltransferase activity / RNA endonuclease activity producing 3'-phosphomonoesters, hydrolytic mechanism / 5'-3' RNA helicase activity / Lyases; Phosphorus-oxygen lyases / mRNA guanylyltransferase activity / Assembly of the SARS-CoV-2 Replication-Transcription Complex (RTC) / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of TBK1 activity / Maturation of replicase proteins / TRAF3-dependent IRF activation pathway / ISG15-specific peptidase activity ...protein guanylyltransferase activity / RNA endonuclease activity producing 3'-phosphomonoesters, hydrolytic mechanism / 5'-3' RNA helicase activity / Lyases; Phosphorus-oxygen lyases / mRNA guanylyltransferase activity / Assembly of the SARS-CoV-2 Replication-Transcription Complex (RTC) / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of TBK1 activity / Maturation of replicase proteins / TRAF3-dependent IRF activation pathway / ISG15-specific peptidase activity / Transcription of SARS-CoV-2 sgRNAs / snRNP Assembly / Translation of Replicase and Assembly of the Replication Transcription Complex / Replication of the SARS-CoV-2 genome / Hydrolases; Acting on ester bonds; Exoribonucleases producing 5'-phosphomonoesters / double membrane vesicle viral factory outer membrane / SARS coronavirus main proteinase / host cell endoplasmic reticulum-Golgi intermediate compartment / host cell endosome / 3'-5'-RNA exonuclease activity / 5'-3' DNA helicase activity / symbiont-mediated degradation of host mRNA / mRNA guanylyltransferase / symbiont-mediated suppression of host ISG15-protein conjugation / symbiont-mediated suppression of host toll-like receptor signaling pathway / G-quadruplex RNA binding / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of IRF3 activity / omega peptidase activity / mRNA (guanine-N7)-methyltransferase / methyltransferase cap1 / SARS-CoV-2 modulates host translation machinery / symbiont-mediated suppression of host NF-kappaB cascade / host cell Golgi apparatus / DNA helicase / symbiont-mediated perturbation of host ubiquitin-like protein modification / methyltransferase cap1 activity / ubiquitinyl hydrolase 1 / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / cysteine-type deubiquitinase activity / Hydrolases; Acting on peptide bonds (peptidases); Cysteine endopeptidases / lyase activity / single-stranded RNA binding / viral protein processing / host cell perinuclear region of cytoplasm / host cell endoplasmic reticulum membrane / RNA helicase / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / copper ion binding / symbiont-mediated suppression of host gene expression / symbiont-mediated activation of host autophagy / viral translational frameshifting / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / lipid binding / host cell nucleus / SARS-CoV-2 activates/modulates innate and adaptive immune responses / ATP hydrolysis activity / proteolysis / RNA binding / zinc ion binding / ATP binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.57 Å | |||||||||
 Authors Authors | Li Z / Yin W | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2021 Journal: Nat Struct Mol Biol / Year: 2021Title: Structural basis for inhibition of the SARS-CoV-2 RNA polymerase by suramin. Authors: Wanchao Yin / Xiaodong Luan / Zhihai Li / Ziwei Zhou / Qingxing Wang / Minqi Gao / Xiaoxi Wang / Fulai Zhou / Jingjing Shi / Erli You / Mingliang Liu / Qingxia Wang / Yi Jiang / Hualiang ...Authors: Wanchao Yin / Xiaodong Luan / Zhihai Li / Ziwei Zhou / Qingxing Wang / Minqi Gao / Xiaoxi Wang / Fulai Zhou / Jingjing Shi / Erli You / Mingliang Liu / Qingxia Wang / Yi Jiang / Hualiang Jiang / Gengfu Xiao / Leike Zhang / Xuekui Yu / Shuyang Zhang / H Eric Xu /  Abstract: The COVID-19 pandemic caused by nonstop infections of SARS-CoV-2 has continued to ravage many countries worldwide. Here we report that suramin, a 100-year-old drug, is a potent inhibitor of the SARS- ...The COVID-19 pandemic caused by nonstop infections of SARS-CoV-2 has continued to ravage many countries worldwide. Here we report that suramin, a 100-year-old drug, is a potent inhibitor of the SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) and acts by blocking the binding of RNA to the enzyme. In biochemical assays, suramin and its derivatives are at least 20-fold more potent than remdesivir, the currently approved nucleotide drug for treatment of COVID-19. The 2.6 Å cryo-electron microscopy structure of the viral RdRp bound to suramin reveals two binding sites. One site directly blocks the binding of the RNA template strand and the other site clashes with the RNA primer strand near the RdRp catalytic site, thus inhibiting RdRp activity. Suramin blocks viral replication in Vero E6 cells, although the reasons underlying this effect are likely various. Our results provide a structural mechanism for a nonnucleotide inhibitor of the SARS-CoV-2 RdRp. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30572.map.gz emd_30572.map.gz | 60.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30572-v30.xml emd-30572-v30.xml emd-30572.xml emd-30572.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30572.png emd_30572.png | 29 KB | ||
| Filedesc metadata |  emd-30572.cif.gz emd-30572.cif.gz | 6.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30572 http://ftp.pdbj.org/pub/emdb/structures/EMD-30572 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30572 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30572 | HTTPS FTP |
-Related structure data
| Related structure data |  7d4fMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30572.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30572.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.069 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : COVID-19 RdRp complex bound to suramin
| Entire | Name: COVID-19 RdRp complex bound to suramin |
|---|---|
| Components |
|
-Supramolecule #1: COVID-19 RdRp complex bound to suramin
| Supramolecule | Name: COVID-19 RdRp complex bound to suramin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Non-structural protein 8
| Macromolecule | Name: Non-structural protein 8 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.034242 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAIASEFSSL PSYAAFATAQ EAYEQAVANG DSEVVLKKLK KSLNVAKSEF DRDAAMQRKL EKMADQAMTQ MYKQARSEDK RAKVTSAMQ TMLFTMLRKL DNDALNNIIN NARDGCVPLN IIPLTTAAKL MVVIPDYNTY KNTCDGTTFT YASALWEIQQ V VDADSKIV ...String: MAIASEFSSL PSYAAFATAQ EAYEQAVANG DSEVVLKKLK KSLNVAKSEF DRDAAMQRKL EKMADQAMTQ MYKQARSEDK RAKVTSAMQ TMLFTMLRKL DNDALNNIIN NARDGCVPLN IIPLTTAAKL MVVIPDYNTY KNTCDGTTFT YASALWEIQQ V VDADSKIV QLSEISMDNS PNLAWPLIVT ALRANSAVKL Q UniProtKB: Replicase polyprotein 1ab |
-Macromolecule #2: Non-structural protein 7
| Macromolecule | Name: Non-structural protein 7 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 9.38 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSKMSDVKCT SVVLLSVLQQ LRVESSSKLW AQCVQLHNDI LLAKDTTEAF EKMVSLLSVL LSMQGAVDIN KLCEEMLDNR ATLQ UniProtKB: Replicase polyprotein 1ab |
-Macromolecule #3: RNA-directed RNA polymerase
| Macromolecule | Name: RNA-directed RNA polymerase / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 107.96525 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSADAQSFLN RVCGVSAARL TPCGTGTSTD VVYRAFDIYN DKVAGFAKFL KTNCCRFQEK DEDDNLIDSY FVVKRHTFSN YQHEETIYN LLKDCPAVAK HDFFKFRIDG DMVPHISRQR LTKYTMADLV YALRHFDEGN CDTLKEILVT YNCCDDDYFN K KDWYDFVE ...String: MSADAQSFLN RVCGVSAARL TPCGTGTSTD VVYRAFDIYN DKVAGFAKFL KTNCCRFQEK DEDDNLIDSY FVVKRHTFSN YQHEETIYN LLKDCPAVAK HDFFKFRIDG DMVPHISRQR LTKYTMADLV YALRHFDEGN CDTLKEILVT YNCCDDDYFN K KDWYDFVE NPDILRVYAN LGERVRQALL KTVQFCDAMR NAGIVGVLTL DNQDLNGNWY DFGDFIQTTP GSGVPVVDSY YS LLMPILT LTRALTAESH VDTDLTKPYI KWDLLKYDFT EERLKLFDRY FKYWDQTYHP NCVNCLDDRC ILHCANFNVL FST VFPPTS FGPLVRKIFV DGVPFVVSTG YHFRELGVVH NQDVNLHSSR LSFKELLVYA ADPAMHAASG NLLLDKRTTC FSVA ALTNN VAFQTVKPGN FNKDFYDFAV SKGFFKEGSS VELKHFFFAQ DGNAAISDYD YYRYNLPTMC DIRQLLFVVE VVDKY FDCY DGGCINANQV IVNNLDKSAG FPFNKWGKAR LYYDSMSYED QDALFAYTKR NVIPTITQMN LKYAISAKNR ARTVAG VSI CSTMTNRQFH QKLLKSIAAT RGATVVIGTS KFYGGWHNML KTVYSDVENP HLMGWDYPKC DRAMPNMLRI MASLVLA RK HTTCCSLSHR FYRLANECAQ VLSEMVMCGG SLYVKPGGTS SGDATTAYAN SVFNICQAVT ANVNALLSTD GNKIADKY V RNLQHRLYEC LYRNRDVDTD FVNEFYAYLR KHFSMMILSD DAVVCFNSTY ASQGLVASIK NFKSVLYYQN NVFMSEAKC WTETDLTKGP HEFCSQHTML VKQGDDYVYL PYPDPSRILG AGCFVDDIVK TDGTLMIERF VSLAIDAYPL TKHPNQEYAD VFHLYLQYI RKLHDELTGH MLDMYSVMLT NDNTSRYWEP EFYEAMYTPH TVLQGGSENL YFQG UniProtKB: Replicase polyprotein 1ab |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #5: 8-(3-(3-aminobenzamido)-4-methylbenzamido)naphthalene-1,3,5-trisu...
| Macromolecule | Name: 8-(3-(3-aminobenzamido)-4-methylbenzamido)naphthalene-1,3,5-trisulfonic acid type: ligand / ID: 5 / Number of copies: 2 / Formula: H3U |
|---|---|
| Molecular weight | Theoretical: 635.643 Da |
| Chemical component information | 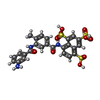 ChemComp-H3U: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 68.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















