+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30299 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Cas12f1-sgRNA-target DNA complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cas12f / Cas14 / sgRNA / target DNA / CRISPR / RNA BINDING PROTEIN-RNA-DNA complex | |||||||||
| Function / homology | Transposase IS605, OrfB, C-terminal / Cas12f1-like, TNB domain / endonuclease activity / defense response to virus / Hydrolases; Acting on ester bonds / DNA binding / RNA binding / metal ion binding / CRISPR-associated endodeoxyribonuclease Cas12f1 Function and homology information Function and homology information | |||||||||
| Biological species |  uncultured archaeon (environmental samples) / synthetic construct (others) uncultured archaeon (environmental samples) / synthetic construct (others) | |||||||||
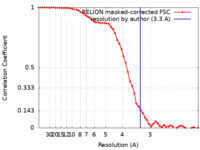
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Takeda NS / Nakagawa R | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2021 Journal: Mol Cell / Year: 2021Title: Structure of the miniature type V-F CRISPR-Cas effector enzyme. Authors: Satoru N Takeda / Ryoya Nakagawa / Sae Okazaki / Hisato Hirano / Kan Kobayashi / Tsukasa Kusakizako / Tomohiro Nishizawa / Keitaro Yamashita / Hiroshi Nishimasu / Osamu Nureki /  Abstract: RNA-guided DNA endonucleases derived from CRISPR-Cas adaptive immune systems are widely used as powerful genome-engineering tools. Among the diverse CRISPR-Cas nucleases, the type V-F Cas12f (also ...RNA-guided DNA endonucleases derived from CRISPR-Cas adaptive immune systems are widely used as powerful genome-engineering tools. Among the diverse CRISPR-Cas nucleases, the type V-F Cas12f (also known as Cas14) proteins are exceptionally compact and associate with a guide RNA to cleave single- and double-stranded DNA targets. Here, we report the cryo-electron microscopy structure of Cas12f1 (also known as Cas14a) in complex with a guide RNA and its target DNA. Unexpectedly, the structure revealed that two Cas12f1 molecules assemble with the single guide RNA to recognize the double-stranded DNA target. Each Cas12f1 protomer adopts a different conformation and plays distinct roles in nucleic acid recognition and DNA cleavage, thereby explaining how the miniature Cas12f1 enzyme achieves RNA-guided DNA cleavage as an "asymmetric homodimer." Our findings augment the mechanistic understanding of diverse CRISPR-Cas nucleases and provide a framework for the development of compact genome-engineering tools critical for therapeutic genome editing. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30299.map.gz emd_30299.map.gz | 2.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30299-v30.xml emd-30299-v30.xml emd-30299.xml emd-30299.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30299_fsc.xml emd_30299_fsc.xml | 5.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_30299.png emd_30299.png | 183.3 KB | ||
| Masks |  emd_30299_msk_1.map emd_30299_msk_1.map | 12.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-30299.cif.gz emd-30299.cif.gz | 6.2 KB | ||
| Others |  emd_30299_half_map_1.map.gz emd_30299_half_map_1.map.gz emd_30299_half_map_2.map.gz emd_30299_half_map_2.map.gz | 9.8 MB 9.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30299 http://ftp.pdbj.org/pub/emdb/structures/EMD-30299 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30299 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30299 | HTTPS FTP |
-Related structure data
| Related structure data |  7c7lMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10585 (Title: Cryo-EM structure of the Cas12f1-sgRNA-target DNA complex EMPIAR-10585 (Title: Cryo-EM structure of the Cas12f1-sgRNA-target DNA complexData size: 649.5 Data #1: Unaligned movies for Cas12f1-sgRNA-target DNA complex [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_30299.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30299.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05133 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_30299_msk_1.map emd_30299_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_30299_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_30299_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of the Cas12f1-sgRNA-target DNA complex
| Entire | Name: Cryo-EM structure of the Cas12f1-sgRNA-target DNA complex |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of the Cas12f1-sgRNA-target DNA complex
| Supramolecule | Name: Cryo-EM structure of the Cas12f1-sgRNA-target DNA complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  uncultured archaeon (environmental samples) uncultured archaeon (environmental samples) |
-Supramolecule #2: Cas12f1
| Supramolecule | Name: Cas12f1 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|
-Supramolecule #4: target DNA
| Supramolecule | Name: target DNA / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #3-#4 |
|---|
-Supramolecule #3: sgRNA
| Supramolecule | Name: sgRNA / type: organelle_or_cellular_component / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|
-Macromolecule #1: CRISPR-associated protein Cas14a.1
| Macromolecule | Name: CRISPR-associated protein Cas14a.1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  uncultured archaeon (environmental samples) uncultured archaeon (environmental samples) |
| Molecular weight | Theoretical: 62.748598 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGHHHHHHGS MAKNTITKTL KLRIVRPYNS AEVEKIVADE KNNREKIALE KNKDKVKEAC SKHLKVAAYC TTQVERNACL FCKARKLDD KFYQKLRGQF PDAVFWQEIS EIFRQLQKQA AEIYNQSLIE LYYEIFIKGK GIANASSVEH YLSDVCYTRA A ELFKNAAI ...String: MGHHHHHHGS MAKNTITKTL KLRIVRPYNS AEVEKIVADE KNNREKIALE KNKDKVKEAC SKHLKVAAYC TTQVERNACL FCKARKLDD KFYQKLRGQF PDAVFWQEIS EIFRQLQKQA AEIYNQSLIE LYYEIFIKGK GIANASSVEH YLSDVCYTRA A ELFKNAAI ASGLRSKIKS NFRLKELKNM KSGLPTTKSD NFPIPLVKQK GGQYTGFEIS NHNSDFIIKI PFGRWQVKKE ID KYRPWEK FDFEQVQKSP KPISLLLSTQ RRKRNKGWSK DEGTEAEIKK VMNGDYQTSY IEVKRGSKIG EKSAWMLNLS IDV PKIDKG VDPSIIGGIA VGVKSPLVCA INNAFSRYSI SDNDLFHFNK KMFARRRILL KKNRHKRAGH GAKNKLKPIT ILTE KSERF RKKLIERWAC EIADFFIKNK VGTVQMENLE SMKRKEDSYF NIRLRGFWPY AEMQNKIEFK LKQYGIEIRK VAPNN TSKT CSKCGHLNNY FNFEYRKKNK FPHFKCEKCN FKENADYNAA LNISNPKLKS TKEEP UniProtKB: CRISPR-associated endodeoxyribonuclease Cas12f1 |
-Macromolecule #2: sgRNA
| Macromolecule | Name: sgRNA / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  uncultured archaeon (environmental samples) uncultured archaeon (environmental samples) |
| Molecular weight | Theoretical: 58.133543 KDa |
| Sequence | String: UUCACUGAUA AAGUGGAGAA CCGCUUCACC AAAAGCUGUC CCUUAGGGGA UUAGAACUUG AGUGAAGGUG GGCUGCUUGC AUCAGCCUA AUGUCGAGAA GUGCUUUCUU CGGAAAGUAA CCCUCGAAAC AAAUUCAUUU GAAAGAAUGA AGGAAUGCAA C GGAAAUUA GGUGCGCUUG GC |
-Macromolecule #3: DNA (40-mer)
| Macromolecule | Name: DNA (40-mer) / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 12.297954 KDa |
| Sequence | String: (DG)(DA)(DA)(DT)(DG)(DG)(DT)(DT)(DG)(DC) (DC)(DA)(DA)(DG)(DC)(DG)(DC)(DA)(DC)(DC) (DT)(DA)(DA)(DT)(DT)(DT)(DC)(DC)(DC) (DA)(DA)(DA)(DT)(DT)(DA)(DG)(DA)(DA)(DA) (DA) |
-Macromolecule #4: DNA (40-mer)
| Macromolecule | Name: DNA (40-mer) / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 12.323922 KDa |
| Sequence | String: (DT)(DT)(DT)(DT)(DC)(DT)(DA)(DA)(DT)(DT) (DT)(DG)(DG)(DG)(DA)(DA)(DA)(DT)(DT)(DA) (DG)(DG)(DT)(DG)(DC)(DG)(DC)(DT)(DT) (DG)(DG)(DC)(DA)(DA)(DC)(DC)(DA)(DT)(DT) (DC) |
-Macromolecule #5: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 5 / Number of copies: 3 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 48.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)