[English] 日本語
 Yorodumi
Yorodumi- EMDB-30194: Cryo-EM structure of Dengue virus serotype 2 complexed with Fab S... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30194 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
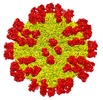
| Title | Cryo-EM structure of Dengue virus serotype 2 complexed with Fab SIgN-3C at pH 6.5 | |||||||||
 Map data Map data | Cryo-EM structure of Dengue virus serotype 2 complexed with Fab SIgN-3C at pH 6.5 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | antibody / neutralization / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host JAK-STAT cascade via inhibition of host TYK2 activity / host cell mitochondrion / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT2 activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / ribonucleoside triphosphate phosphatase activity / double-stranded RNA binding / channel activity / viral capsid / monoatomic ion transmembrane transport / clathrin-dependent endocytosis of virus by host cell ...symbiont-mediated suppression of host JAK-STAT cascade via inhibition of host TYK2 activity / host cell mitochondrion / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT2 activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / ribonucleoside triphosphate phosphatase activity / double-stranded RNA binding / channel activity / viral capsid / monoatomic ion transmembrane transport / clathrin-dependent endocytosis of virus by host cell / mRNA (nucleoside-2'-O-)-methyltransferase activity / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / RNA helicase activity / protein dimerization activity / host cell endoplasmic reticulum membrane / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / induction by virus of host autophagy / viral RNA genome replication / serine-type endopeptidase activity / RNA-dependent RNA polymerase activity / virus-mediated perturbation of host defense response / fusion of virus membrane with host endosome membrane / viral envelope / host cell nucleus / virion attachment to host cell / structural molecule activity / virion membrane / proteolysis / extracellular region / ATP binding / membrane / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Dengue virus 2 Dengue virus 2 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Zhang S / Chew SV | |||||||||
| Funding support |  Singapore, 2 items Singapore, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2020 Journal: Cell Rep / Year: 2020Title: A Human Antibody Neutralizes Different Flaviviruses by Using Different Mechanisms. Authors: Shuijun Zhang / Thomas Loy / Thiam-Seng Ng / Xin-Ni Lim / Shyn-Yun Valerie Chew / Ter Yong Tan / Meihui Xu / Victor A Kostyuchenko / Farhana Tukijan / Jian Shi / Katja Fink / Shee-Mei Lok /  Abstract: Human antibody SIgN-3C neutralizes dengue virus (DENV) and Zika virus (ZIKV) differently. DENV:SIgN-3C Fab and ZIKV:SIgN-3C Fab cryoelectron microscopy (cryo-EM) complex structures show Fabs ...Human antibody SIgN-3C neutralizes dengue virus (DENV) and Zika virus (ZIKV) differently. DENV:SIgN-3C Fab and ZIKV:SIgN-3C Fab cryoelectron microscopy (cryo-EM) complex structures show Fabs crosslink E protein dimers at extracellular pH 8.0 condition and also when further incubated at acidic endosomal conditions (pH 8.0-6.5). We observe Fab binding to DENV (pH 8.0-5.0) prevents virus fusion, and the number of bound Fabs increase (from 120 to 180). For ZIKV, although there are already 180 copies of Fab at pH 8.0, virus structural changes at pH 5.0 are not inhibited. The immunoglobulin G (IgG):DENV structure at pH 8.0 shows both Fab arms bind to epitopes around the 2-fold vertex. On ZIKV, an additional Fab around the 5-fold vertex at pH 8.0 suggests one IgG arm would engage with an epitope, although the other may bind to other viruses, causing aggregation. For DENV2 at pH 5.0, a similar scenario would occur, suggesting DENV2:IgG complex would aggregate in the endosome. Hence, a single antibody employs different neutralization mechanisms against different flaviviruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30194.map.gz emd_30194.map.gz | 267.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30194-v30.xml emd-30194-v30.xml emd-30194.xml emd-30194.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30194.png emd_30194.png | 271 KB | ||
| Filedesc metadata |  emd-30194.cif.gz emd-30194.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30194 http://ftp.pdbj.org/pub/emdb/structures/EMD-30194 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30194 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30194 | HTTPS FTP |
-Validation report
| Summary document |  emd_30194_validation.pdf.gz emd_30194_validation.pdf.gz | 769.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30194_full_validation.pdf.gz emd_30194_full_validation.pdf.gz | 769.4 KB | Display | |
| Data in XML |  emd_30194_validation.xml.gz emd_30194_validation.xml.gz | 7.7 KB | Display | |
| Data in CIF |  emd_30194_validation.cif.gz emd_30194_validation.cif.gz | 8.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30194 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30194 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30194 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30194 | HTTPS FTP |
-Related structure data
| Related structure data |  7bubMC  7bu8C  7buaC  7budC  7bueC  7bufC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30194.map.gz / Format: CCP4 / Size: 290.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30194.map.gz / Format: CCP4 / Size: 290.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of Dengue virus serotype 2 complexed with Fab SIgN-3C at pH 6.5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.7 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of Dengue virus serotype 2 with SIgN-3C Fab
| Entire | Name: Complex of Dengue virus serotype 2 with SIgN-3C Fab |
|---|---|
| Components |
|
-Supramolecule #1: Complex of Dengue virus serotype 2 with SIgN-3C Fab
| Supramolecule | Name: Complex of Dengue virus serotype 2 with SIgN-3C Fab / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #2: SIgN-3C Fab
| Supramolecule | Name: SIgN-3C Fab / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Dengue virus 2 Dengue virus 2 |
-Supramolecule #3: Dengue virus serotype 2
| Supramolecule | Name: Dengue virus serotype 2 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#4 |
|---|
-Macromolecule #1: SIgN-3C Fab heavy chain
| Macromolecule | Name: SIgN-3C Fab heavy chain / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.448958 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVQSGPD VEKPGASVKV SCKASGYTFT SNYIHWVRQA PGQGLEWMGV INPRGGSTAS AQKFQGRITM TRDTSTSTVY MELSSLRSD DTAVYYCARG GRALFYDSYT TPRDGGSWWF DPWGQGSLVT VSS |
-Macromolecule #2: SIgN-3C Fab light chain
| Macromolecule | Name: SIgN-3C Fab light chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.832145 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQLTQSPSS LSASVGDRVT FTCQASQDIR KYLNWYQQKP GKAPKLLIYD ASNLKTGVPS RFSGSGSGTD FTFTISSLQP EDVATYYCQ QFDDLPITFG QGTRLQIK |
-Macromolecule #3: Dengue virus serotype2 E protein
| Macromolecule | Name: Dengue virus serotype2 E protein / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Dengue virus 2 Dengue virus 2 |
| Molecular weight | Theoretical: 54.258551 KDa |
| Sequence | String: MRCIGISNRD FVEGVSGGSW VDIVLEHGSC VTTMAKNKPT LDFELIKTEA KHPATLRKYC VEAKLTNTTT ASRCPTQGEP SLNEEQDKR FVCKHSMVDR GWGNGCGLFG KGGIVTCAMF TCKKNMEGKV VQPENLEYTI VITPHSGEEN AVGNDTGKHG K EIKVTPQS ...String: MRCIGISNRD FVEGVSGGSW VDIVLEHGSC VTTMAKNKPT LDFELIKTEA KHPATLRKYC VEAKLTNTTT ASRCPTQGEP SLNEEQDKR FVCKHSMVDR GWGNGCGLFG KGGIVTCAMF TCKKNMEGKV VQPENLEYTI VITPHSGEEN AVGNDTGKHG K EIKVTPQS SITEAELTGY GTVTMECSPR TGLDFNEMVL LQMENKAWLV HRQWFLDLPL PWLPGADTQG SNWIQKETLV TF KNPHAKK QDVVVLGSQE GAMHTALTGA TEIQMSSGNL LFTGHLKCRL RMDKLQLKGM SYSMCTGKFK VVKEIAETQH GTI VIRVQY EGDGSPCKIP FEIMDLEKRH VLGRLITVNP IVTEKDSPVN IEAEPPFGDS YIIIGVEPGQ LKLSWFKKGS SIGQ MFETT MRGAKRMAIL GDTAWDFGSL GGVFTSIGKA LHQVFGAIYG AAFSGVSWTM KILIGVVITW IGMNSRSTSL SVSLV LVGV VTLYLGVMVQ A |
-Macromolecule #4: Dengue virus serotype 2 M protein
| Macromolecule | Name: Dengue virus serotype 2 M protein / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Dengue virus 2 Dengue virus 2 |
| Molecular weight | Theoretical: 8.109491 KDa |
| Sequence | String: SVALVPHVGM GLETRTETWM SSEGAWKHAQ RIETWILRHP GFTIMAAILA YTIGTTYFQR VLIFILLTAV TP |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.2 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 16300 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















