+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30017 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | AtMSL1 wild type | |||||||||
 Map data Map data | EM map at 3.3 angstrom | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mechanosensitive / Ion channel / Plant / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationchloroplast envelope / mechanosensitive monoatomic ion channel activity / chloroplast / cellular response to oxidative stress / mitochondrial inner membrane / mitochondrion Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Sun L | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2020 Journal: Cell Rep / Year: 2020Title: Structural Insights into a Plant Mechanosensitive Ion Channel MSL1. Authors: Yawen Li / Yufei Hu / Jiawei Wang / Xin Liu / Wei Zhang / Linfeng Sun /  Abstract: The small conductance mechanosensitive ion channel (MscS)-like (MSL) proteins in plants are evolutionarily conserved homologs of the bacterial small conductance mechanosensitive ion channels. As the ...The small conductance mechanosensitive ion channel (MscS)-like (MSL) proteins in plants are evolutionarily conserved homologs of the bacterial small conductance mechanosensitive ion channels. As the sole member of the Arabidopsis MSL family localized in the mitochondrial inner membrane, MSL1 is essential to maintain the normal membrane potential of mitochondria. Here, we report a cryoelectron microscopy (cryo-EM) structure of Arabidopsis thaliana MSL1 (AtMSL1) at 3.3 Å. The overall architecture of AtMSL1 is similar to MscS. However, the transmembrane domain of AtMSL1 is larger. Structural differences are observed in both the transmembrane and the matrix domain of AtMSL1. The carboxyl-terminus of AtMSL1 is more flexible and the β-barrel structure observed in MscS is absent. The side portals in AtMSL1 are significantly smaller, and enlarging the size of the portal by mutagenesis can increase the channel conductance. Our study provides a framework for eukaryotic MscS-like mechanosensitive ion channels and the gating mechanism of the MscS family. | |||||||||
| History |
|
- Structure visualization
Structure visualization
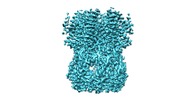
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30017.map.gz emd_30017.map.gz | 49.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30017-v30.xml emd-30017-v30.xml emd-30017.xml emd-30017.xml | 13.8 KB 13.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30017.png emd_30017.png | 238.7 KB | ||
| Filedesc metadata |  emd-30017.cif.gz emd-30017.cif.gz | 5.3 KB | ||
| Others |  emd_30017_half_map_1.map.gz emd_30017_half_map_1.map.gz emd_30017_half_map_2.map.gz emd_30017_half_map_2.map.gz | 40.5 MB 40.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30017 http://ftp.pdbj.org/pub/emdb/structures/EMD-30017 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30017 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30017 | HTTPS FTP |
-Related structure data
| Related structure data |  6lypMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30017.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30017.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM map at 3.3 angstrom | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Half 1 map
| File | emd_30017_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half 1 map | ||||||||||||
| Projections & Slices |
| ||||||||||||
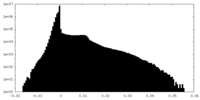
| Density Histograms |
-Half map: Half 2 map
| File | emd_30017_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half 2 map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Wild type of AtMSL1
| Entire | Name: Wild type of AtMSL1 |
|---|---|
| Components |
|
-Supramolecule #1: Wild type of AtMSL1
| Supramolecule | Name: Wild type of AtMSL1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 350 KDa |
-Macromolecule #1: Mechanosensitive ion channel protein 1, mitochondrial
| Macromolecule | Name: Mechanosensitive ion channel protein 1, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 53.942 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAGVRLSLLK SIQRSIKPHA TAKSCSGLLN SHARAFTCGN LLDGPKASPS MISFSSNIRL HNDAKPFNYL GHSSYARAFS SKSDDFGSI VASGVTGSGD GNGNGNDWVE KAKDVLQTSV DAVTETAKKT KDVSDEMIPH VQQFLDSNPY LKDVIVPVSL T MTGTLFAW ...String: MAGVRLSLLK SIQRSIKPHA TAKSCSGLLN SHARAFTCGN LLDGPKASPS MISFSSNIRL HNDAKPFNYL GHSSYARAFS SKSDDFGSI VASGVTGSGD GNGNGNDWVE KAKDVLQTSV DAVTETAKKT KDVSDEMIPH VQQFLDSNPY LKDVIVPVSL T MTGTLFAW VVMPRILRRF HTYAMQSSAK LLPVGFSNED VPYEKSFWGA LEDPARYLVT FIAFAQIAAM VAPTTAIAQY FS PTVKGAV ILSLVWFLYR WKTNVITRML SAKSFGGLDR EKVLTLDKVS SVGLFAIGLM ASAEACGVAV QSILTVGGVG GVA TAFAAR DILGNVLSGL SMQFSRPFSM GDTIKAGSVE GQVIEMGLTT TSLLNAEKFP VLVPNSLFSS QVIVNKSRAQ WRAI ASKIP LQIDDLDMIP QISNEIKEML RSNTKVFLGK EAPHCYLSRV EKSFAELTIG CNLIRMGKEE LYNTQQEVLL EAVKI IKKH GVSLGTTWDN STL UniProtKB: Mechanosensitive ion channel protein 1, mitochondrial |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C7 (7 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 3.3 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.1) / Number images used: 251528 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)